Role and Molecular Mechanisms of Lysosomes and Cathepsins in Neuropathology and Aging: New Insights
Novelty of the Study
Lysosomes are the major catabolic machinery that degrades several biomolecules and pathogens. They do so by a vast assortment of their cathepsins and help in maintaining homeostasis. Cathepsins and their variants are reported to be involved in multiple physiological activities including apoptosis, coagulation, cell proliferation, immunity, and digestion, etc. Here, we have reviewed the role of lysosomes and cathepsins in various nervous system pathologies and aging. Moreover, we have provided an insightful approach that targeting these cathepsins may promise better future therapies to improve not only the life span but also the healthspan.
Abstract
Lysosomes are the major catabolic entities that target a large number of biomolecules and pathogens as well. These cytoplasmic organelles play a substantial role in maintaining homeostasis with the aid of a vast assortment of its cathepsins. Cathepsins and its variants are reported to be involved in multiple physiological activities including apoptosis, coagulation, cell proliferation, immunity, and digestion, etc. These cathepsins are responsible for apoptosis leading to neuropathological changes and aging. Among various types of cathepsins, cathepsin B, D and L, are responsible for aging and neurodegenerative disorders, either by activation of microglia or production of inflammosomes. Aging is also attributed to lysosomal storage diseases caused by impairment in lysosomal function and lipofuscin accumulation. So, lysosomes are not only the dumping ground of the cellular waste but also tightly linked to autophagy at the signaling level. This balance has to be strictly controlled by catabolic capacity of the cell, which is accomplished by ubiquination and autophagic machinery to control aging. Furthermore, lysosomes play critical role in life expectancy via a vast variety of signaling pathways and enhanced lysosmal activity that can improve life span. Taken together these considerations, this review provides an insight into the role of cathepsins in various animal models in autophagy and aging. Furthermore, molecular mechanisms underlying the role of lysosomes and altered expression of cathepsins in neurodegenerative changes and cellular longevity has been reported. Thus, improvement or enhancement in lysosomal activity may prove an ideal strategy to extend life span.
Keywords
Aging, Brain, Cathepsins, Healthspan, Life span, Lysosomes, Neurodegeneration, Neurons
Introduction
Lysosomes, single membrane-bounded organelles, are characterized as major catabolic entities and have been reported in all types of animal cells except red blood cells (RBCs). There is a long list of substances targeted and degraded by lysosomes including pathogens, a variety of biomolecules, surface proteins, old/worn-out organelles, and many other particles. Catabolic function of lysosome is attributed to a variety of acidic hydrolases. Lysosomes play their substantial role in a variety of biological activities like development, apoptosis, waste disposal, plasma membrane repair and maintenance, stress management, nutrient supply, cellular differentiation, and many more to help and maintain cellular homeostasis [1-3]. Discovery of lysosomes was a milestone achievement to get an insight into proteolysis [4]. They have a battery of enzymes and also named as suicide bags because of their involvement in the intracellular degradation of different substances. Lysosomal hydrolases also have a very specific role in protein degradation [5].
Cathepsins are a group of protein-digesting enzymes and are classified into various families i.e. serine, cysteine and aspartyl proteases [6]. Fifteen (15) classes of cathepsins in total have been identified so far [7]. These lysosomal enzymes are highly dynamic in their actions and are involved in multiple physiologic activities i.e. autophagy, apoptotic cell death, blood coagulation, cell proliferation, immunity, and digestion among many more [8-10].
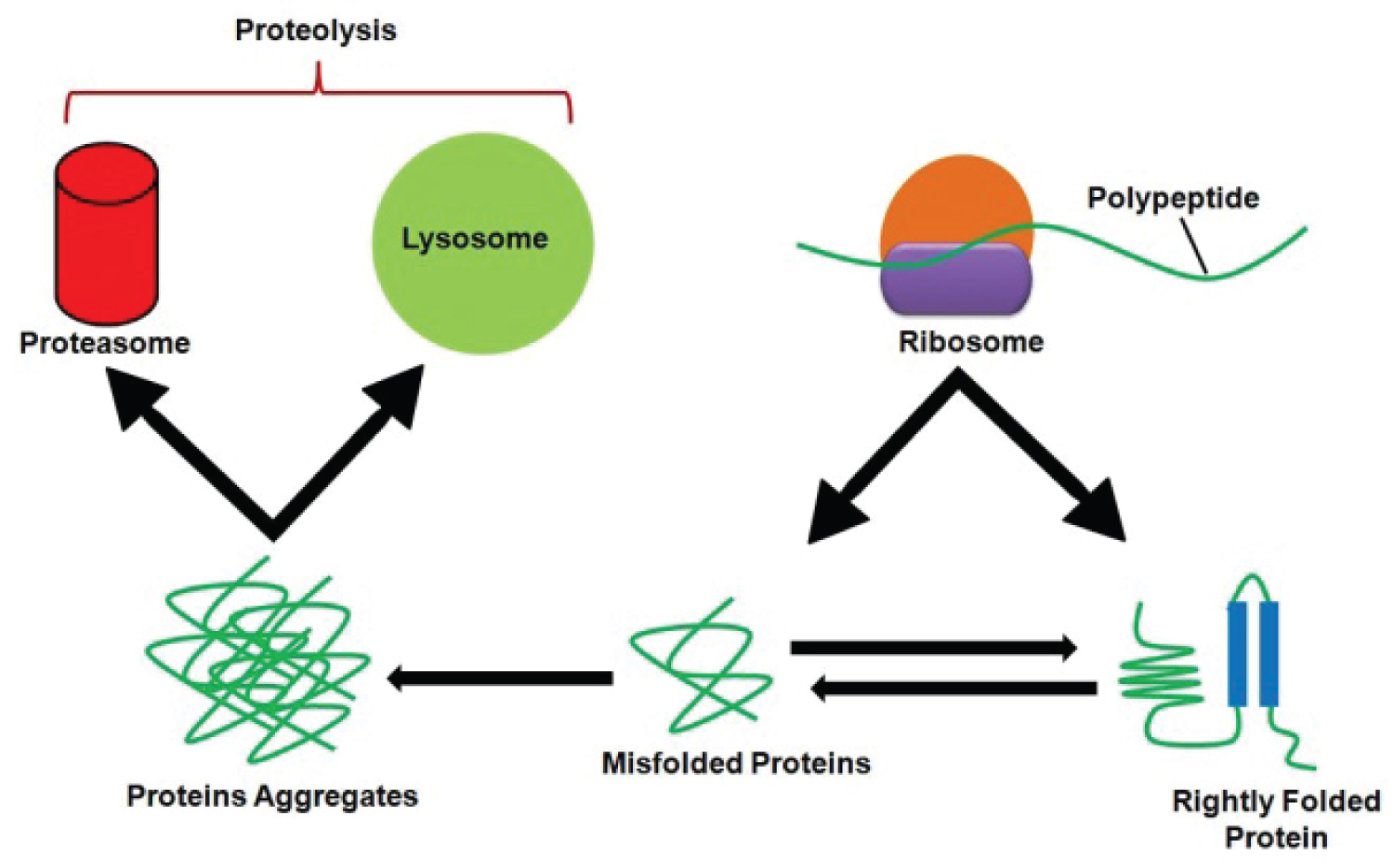
Proteins are continuously being synthesized and degraded in the cell [11]. This is a time saving strategy of the proteins during destruction or alteration when exposed to an unsafe cellular environment. Superficially, the renewal of cellular components seems to be a waste of resources but actually, it is beneficial to the cell because the accumulation of futile components may jeopardize the survival and function of the cell [12]. Besides, protein breakdown is a recycling process as the resulting amino acids are reused [11]. However, the rate of protein degradation varies among cells, under different circumstances and in response to different stimuli. The balance between protein breakdown and protein synthesis is very crucial as it helps the cell to adapt always changing extracellular surroundings by rapidly modifying intracellular proteins. The cell responds to stress routinely by activating its protein degradation systems [12,13]. The role of these proteolytic systems in the well-being of the cell is supported by molecular chaperones. These chaperones determine whether to refold, repair or destroy the proteins. If suitable, they help to repair or refold the proteins, otherwise, proteins are destined to proteolysis (Figure 1). Proteolysis is a fundamental requirement in the elimination of pathogens/invaders and some developmental processes i.e. differentiation, morphogenesis, and embryogenesis [14,15]. There are two key systems for protein breakdown: Rapid non-lysosomal protein degradation system or ubiquitin-proteasome system [16] and the second one is the lysosome-autophagy system [13,16].
Aging and Lysosomes
Aging is characterized by inevitable poor physiological integrity and altered or impaired body activities thus enhance the liability to death. Nine hallmarks of aging include deregulated nutrient sensing, stem cell exhaustion, genomic instability, lost proteostasis, altered intercellular communication, telomere attrition, mitochondrial dysfunction, cellular senescence, and epigenetic alterations [17]. Furthermore, malfunctioned intracellular proteolytic mechanisms arbitrated by the lysosome results in collapsed homeostasis [18]. Cathepsins trigger apoptosis in aging and impaired endosomal-lysosomal neuron system [19]. Among various types of cathepsins, cathepsins B, D, and L are responsible for aging. Previously, injection of leupeptin (cysteine cathepsin inhibitor) or chloroquine (a lysosomotropic drug) into the brain of young rats led to decreased dopamine receptors and accumulation of lipofuscin which are otherwise observed only in aged brain [20,21]. Cathepsin B is a lysosomal cysteine protease and is known to be involved in neuropathological and neurodegenerative disorders [8,22]. Leakage of cathepsin B from the lysosome into the cytosol in microglia is associated with cognitive impairment [23]. Nakanishi, et al. [24] reported that the level of cathepsin B in the brain of all the aged animals except in the striatum have relatively constant activity, where its activity increases with age. Also, a significantly decreased level of cathepsin L has been observed in the brain of aged rats. The presence of a catalytically inactive form of cathepsin L can be suggested from this study, probably because of lysosomal pH alterations as it is highly susceptible to a high pH [24]. Meganeuritis and propagation of lysosome were also observed in the hippocampus upon cathepsins B and L inhibition suggesting that meganeurite may store catabolic organelles. Separation of these meganeurites from parent cells was proposed to result in axotomy. So, the inhibition of cathepsin L may trigger a faster gerontological sequence somewhat resembling aging in cultured slices [25]. Porta, et al. [26] reported a constant level of cathepsin B histochemically in the brain of 14-24 months-old rats [26]. The presence of cathepsin B has been confirmed immunologically in several neurons and astrocytes [27]. Microglia comprises about 20% of the total glial cell population and are distributed in the central nervous system (CNS). Microglia, once activated, are sources of cathepsins and play a key role in neuroinflammation and neurodegenerative diseases in aging [28-30]. In response to Aβ, NALP3 inflammasome-mediated production of IL-1β is because of cathepsin B [31]. Similarly, the expression of another microglia-secreted cathepsin S has been found enhancing during CNS inflammation and aging in mice [32]. Altered expression of cathepsin is controlled by a built-in molecular clock in cortical microglia, that shows the circadian expression of cathepsin S which is involved in diurnal variations of synaptic strength in these neurons via proteolytic modification. Cathepsin S has also been associated with some sleeping disorders as its genetic ablation results in reduced synaptic strength during sleep, by inducing hyperlocomotor activity, as it is required to get novel information after waking [33]. Moreover, cathepsin X (expressed in immune cells) has also been reported to be upregulated in microglia as well as in brain tissues during aging and associated with neuroinflammation in various models of neurodegenerative diseases [32]. Cathepsin X regulates brain function via gamma enolase and may be involved in Alzheimer's disease [34]. Another cathepsin, C (cysteine cathepsin) is a key cathepsin in neuronal and brain functions. Although, cathepsin C is expressed in low level but still has a significant contribution in neuroinflammatory process accompanying pathogenesis in CNS. Previous studies have described the age-dependent changes in the expression of cathepsin D [35-40]. A significant increase in cathepsin D activity in various parts of the brain i.e. cerebellum, cerebrum, pons, and hippocampus was noted with aging in Wistar rats [39]. Banay-Schwartz, et al. (1992) studied the distribution of the cathepsin D activity in fifty distinct areas of CNS in adult and aged humans with a less pronounced alteration in the activity with advancing age as compared to the rat brain [35]. A promising relationship between cathepsin D activation and senile plaques establishment was proposed in aging and Alzheimer's disease [41]. Contrary to young adult rats, cathepsin E (an aspartic protease) was hardly detected in any brain part. The activity of cathepsin D amplified in aged rats compared to the young rats in all brain tissues, but levels of cathepsin E were found to be increased only in the neostriatum and cerebral cortex [24]. The higher expression of cathepsins D and E and their co-localization with lipofuscin in aged rat's neurons and brain are in complete agreement with a previous study that indicates alterations in the lysosomal proteolytic system, suggesting an altered lipofuscinogenesis and intracellular APP (amyloid precursor protein) metabolism [20,24]. Variation in cathepsin D expression during postnatal development of CNS in the rat is suggestive of a possible role of this important enzyme in myelination [42]. Cathepsin D was found in neurons and glial cells of humans. Its level markedly elevates during neuro-ontogenesis [36]. During the age transition, pronounced alterations were observed in lysosomes of different regions of the rat brain [43]. In 35% of neurons of cerebral cortex age-related changes viz, translocation of cathepsin D translocation from lysosomes to cytosolic granules were observed. However, the pattern of variation of cathepsin B indicates that it regulates brain aging differently. Translocation of cathepsin D from the lysosome to cytosol leads to cell death in the aged brain [44]. Earlier, instability of the lysosome was considered as a reason for the release of cathepsin D [39]. However, the translocation of this enzyme was not due to the changed permeability of the lysosomal membrane. Later on, cathepsin D was anticipated as a potential biomarker of aging [45]. Overexpression and translocation of cathepsin D result in the presence of molecular targets of cathepsins in CNS. Cathepsin D has involved in the breakdown of several proteins i.e. tubulin and microtubule-associated proteins (MAP-1 & MAP-2) in an age-dependent manner [38]. These alterations suggest that substrate and enzyme specificity is vulnerable during aging [46]. In the cytosol of the hippocampus, tau protein serves as a target of cathepsin D; but requires suppression of cathepsin B. This suppression results in an increased lysosomal pH that ultimately enhance the proteolysis of tau protein. During brain aging, the shift of pH may ignite a series of reactions in the inactivation of cathepsin L that in turn stimulates the expression of cathepsin D resulting in proteolysis of tau [25]. There are several factors that affect the aging (Table 1).
Lysosomes are key players in several cellular processes that control organismal death and apoptosis. Thus, any impairment in lysosomal function may lead to several lysosomal storage diseases (LSDs) [47]. Mutations in any of the genes responsible for hydrolases can lead to LSDs with infants being more susceptible than adults but sometimes adults are also affected by LSDs that result in neurodegeneration [19]. Several organs are affected and undergo tissue degeneration, however, LSDs have distinct effects both in patients and animal models [48]. CNS is most vulnerable that is adversely affected by lysosomal dysfunctions [49,50]. Age-related neurodegenerative pathologies i.e. Parkinson's or Alzheimer's diseases have also been linked to lysosomal defects including perturbed lysosomal proteolysis, disturbance in calcium homeostasis and a decreased intraluminal fermentation [51,52]. Malfunctioning of the two key lysosomal enzymes i.e. glucosylceramidase and glucocerebrosidase (GBA) may enhance the risk of Parkinson's disease several times. Similarly, deficiency of GBA may cause an increase in the levels of alpha-synuclein whose atypical accumulation ultimately results in synucleopathies i.e. Parkinson's disease [53]. These kinds of anomalies disturb an intricate balance between the production and proteolytic clearance of damaged or abnormal proteins that may result in massive protein aggregates that can pass to progeny. These massive protein aggregates may results in the formation of lipofuscin by reacting with other bio components i.e. metals and lipids [54]. Lipofuscin can lead to generation of oxidants either by inhibition of proteasome or iron-mediated catalysis of free radicals, thus accumulation of lipofuscin aggregates can further add to neurodegeneration by oxidative stress [55]. As a result, overall wellbeing and life expectancy of the individual are decreased. Considering the wide intra-cellular effects and range of lysosomal functions, aging is characterized by a decreased lysosomal activity at the molecular level. Furthermore, autophagy and lysosomal degradation are closely linked as they help removing superfluous and material accumulated due to aging [56]. So, lysosomes are not only the dumping ground of the cellular waste but also tightly linked to autophagy at the signaling level. Lysosomes are a molecular hub of mTORC1-kinase complex activity that negatively regulates the autophagic process. They are also associated with transcription factor EB (TFEB), a crucial factor in lysosomal biogenesis and an activator of autophagy [1]. In addition to its prime role in autophagy, lysosomes play a critical role in life expectancy via a vast variety of signaling pathways. For example, in worms, active usage of lipids from lysosomes to the nucleus resulted in perturbed gene expression of life expectancy controlling genes [57]. Moreover, in yeast, intra-lysosomal pH influenced the lysosomal storage and degradative activity and suspected to be an active aging-relevant signal. Aging is also significantly controlled and affected by metals (i.e. Ca++ and Fe++) and stored substances (i.e. amino acids), these substances are vital to maintain the coordinated lysosomal expression and regulation (CLEAR) network [58-61].
Lysosomes and Autophagy Regulation in Aging
The survival of an organism is governed by its capability to sustain a balance between the production of new stuff and the degradation of old harmful cellular structures. This balance is strictly controlled by catabolic capacity of the cell, which is accomplished by two processes: 1) Ubiquitin/proteasomal degradation pathway, mainly targets cytosolic and endoplamic reticulum (ER) proteins following their retrograde transport to the cytosol while ubiquitinated mitochondrial proteins are exposed [62,63], 2) Autophagic machinery destroys cytosolic substrates, from single proteins to whole organelles which are delivered to the lysosome for hydrolytic damage. Both pathways control aging [64-67], and also interact with each other [68], but their role in life span control still needs more investigation. Three major subcategories of autophagy are chaperone-mediated autophagy (CMA), macroautophagy, and microautophagy [69]. CMA is a selective protein targeting pathway that directs the substrate translocation to lysosomes during aging [70,71]. Microautophagy involves direct invagination of cytosolic material at the lysosomal membrane [72] which needs further investigations [71]. Macroautophagy is a recyclable process of lysosomes that maintains cellular homeostasis [73]. In macroautophagy, the target material is engulfed in autophagosomes, transported to the lysosome, where the membrane of autophagosome fuses with lysosome causing the formation of a single membrane vesicle which is degraded in the lumen [74]. Macroautophagic machinery is exceedingly conserved, from unicellular yeast to mammals [75]. Macroautophagy is responsible for life span control [56,71,76-78]. The rate of autophagy is decreased in aging [71]. On the contrary, pharmacological and genetic modulations enhance autophagic activity by extending longevity. Thus, aging is controlled by the regulatory genes of all steps of autophagic process viz, 1) Regulation/initiation, 2) Phagophore formation, 3) Cargo loading and degradation [56,78]. Autophagy deficiency has been found to decrease the life span and in contrast, accelerated age-related pathologies [74]. For instance, mutations in any of the two principal initiation complexes of autophagy (ULK1 and Beclin-1) resulted in a reduced life span, premature aging and age-related pathologies [79,80]. Tissue-specific ATG5-KO and ATG7-KO resulted in accelerated aging and age-related problems [49]. These problems include the accumulation of lipofuscin [77], lipid droplets, defective mitochondria [81], and excessive protein oxidation along with neurodegeneration [50,82]. The third step of autophagy can further be divided into three sub-stages: 1) Cargo recognition and loading; 2) Delivery to and fusion with a lysosome; 3) Lysosomal degradation. Any defect in these stages may lead to an abnormal autophagic flux [83] and may result in many neurodegenerative disorders, including Parkinson's disease, Huntington's disease, Alzheimer's disease and amyotrophic lateral sclerosis (ALS). Furthermore, a mutation in p62, which is a crucial marker for autophagic flux especially in the cargo‐recognition machinery which transports substrates to autophagosomes can further enhance autophagy and lead to onset of ALS and Paget's disease [84-86]. These findings suggest that autophagy is commonly aberrant in neurodegenerative diseases. However, further studies are still required to explore specific autophagic or signaling pathways involved in different types or sub‐types of neurodegenerative diseases [87].
Lysosomal contributions in anti-aging activities:
Progressive lysosomal dysfunction can lead to aging, and based on this information a question arises that whether any improvement or enhanced lysosomal activity can improve or extend life span or not? A large number of studies carried out in various model organisms have proven this speculation to be right. For instance, an improvement in life span was observed in S. cerevisiae upon over-expression of Pep4 (a homolog of cathepsin D) [88]. Similarly, an upregulated level of LIPL-4 (lysosomal acid lipase) has been attributed to an extension in the life span of C. elegans [89]; via fatty acid oleoylethanolamide (OEA) production [57]. Furthermore, life span extension via autophagy induction has been observed as a result of overexpression of TFEB homolog in C. elegans (HLH-30) [90] as HLH-30 along with MXL-3 (another transcription factor) is responsible for lysosomal lipolysis. Therefore, a mutation in MXL-3 led to an extension of the life span [91]. Overexpression of N-ethyl-maleimide sensitive fusion protein (NSF1) resisted the development of neurodegeneration in D. melanogaster perhaps via activation of lysosomal proteases and autophagy induction [92]. Likewise, Akt2 ablation prolongs life span and improves myocardial contractile function with a possible adaptive cardiac remodeling through the sirutin 1 (SIRT1) mediated autophagy regulation in mice [93]. Furthermore, induction of autophagy using rapamycin, resveratrol, nicotinamide derivatives, metformin, urolithin A, or spermidine have been reported in previous studies to delay aging, prolong life span, and improve cardiovascular function in aging [94]. Taking together these findings it can be concluded that increased lysosomal performance may protect organisms from aging-associated pathologies. Similarly, the lysosomal function improves healthspan and life span. This association emerges from the important role of lysosomes observed in autophagy [78]. Life span extension is strictly related to the induction of autophagy and therefore controls aging. Spermidine is a naturally occurring polyamine [95] and its level decreases with age and triggers autophagy. Spermidine treatment in yeast brings about histone H3 hypo-acetylation probably by inhibition of acetyltransferases, which results in loss of autophagy essential gene, ATG7 [96]. Autophagy is induced by spermidine injections, accompanied by inhibition of acetylation of different cytosolic proteins [97]. Resveratrol is a polyphenolic compound present in grapes and red wine and affects stress-related targets, among them, is the Nicotinamide adenine dinucleotide (NAD+) dependent deacetylase SIRT1 [98]. Interestingly, SIRT1 stimulation causes the deacetylation of autophagic proteins and their succeeding activation [99]. In C. elegans, autophagy activation is necessary for longevity mediated by resveratrol [100]. Certain anti-aging interventions appear due to the combined effect of direct or indirect repression of target of rapamycin (TOR) signaling [101]. Likewise, rapamycin, a direct mTORC1-inhibitor also act as a strong inducer of autophagy [102], among mTOR-independent pathways, the transient receptor potential (TRP) calcium ion channel TRPML (mucolipin) subfamily is emerging as an important signaling channel to modulate lysosomal biogenesis and autophagy [73]. Similarly, activated acetyltransferase MEC-17 promotes autophagy by stimulating cellular microtubule transport machinery [103]. On the other hand, biguanide metformin secondarily inhibits TOR activity by inhibiting oxidative phosphorylation, which brings about a rise in AMP/ATP ratio and activation of AMPK [104]. In the meantime, metformin can up-regulate REDD1 and hinder Ras-related GTP binding (Rag) GTPases, and together they promote repression of TOR [105]. The contribution of lysosomes in these mediations presumably traverses pass their degradative potential. mTORC1-docking is essential for the activity of lysosomes and lysosomes control their mTORC1-docking on their surface by a luminal cargo of amino acids, essential for its movement [106]. Some novel anti-aging interventions may go to the bleeding edge. Dietary supplementation of lysosomally produced OEA advances life span in worms [57] and can subsequently be a potential contender for the dietary anti-aging approach. An exceptionally successful behavioral attitude in contrast to aging is exercise, which not only increases life span [107] but also has universal health benefits [108]. Exercise has been displayed to begin autophagy flux in the muscle [109]. Interestingly, autophagy induced by exercise entails the release of lysosomal Ca2+ from lysosomal calcium channel mucolipin 1 and succeeding activation of calcineurin which in sequence stimulates nuclear translocation of TFEB [61]. Most of the anti-aging involvements converge in lysosome at different levels, highlighting lysosomal function as a vital molecular hub for controlling health span and life span.
Some other molecular mechanisms which are central to autophagy regulation and impact life span are transcriptional and epigenetic regulation of autophagy genes. Recent studies have shown that tissue-specific induction of autophagy can also result in non-cell autonomous extension of organismal life span [110]. Although, the mechanism of non-cell autonomous autophagic regulation is currently unclear, inter-tissue communication of stress responses such as the heat shock response and unfolded protein responses in the endoplasmic reticulum (UPRER) and mitochondrial (UPRmt) have been demonstrated [111]. These organelle-specific and intracellular proteostatic mechanisms may potentially be involved in depicting autophagic status between tissues to regulate aging. Also, autophagy proteins can also possess some autophagy independent functions [112] such as extracellular protein secretion [113], that may mediate inter-tissue integration of nutrient signalling, metabolism, and gene regulation to modulate proteostasis. As certain temporal and circadian considerations are now being integrated in aging studies [114], the underlying mechanisms that affect autophagy and proteostasis and lead to neurodegeneration are bound to be clarified and new exploitable mechanisms are likely to be uncovered.
Conclusion
Taking into account the above literature it can be concluded that lysosomes are major catabolic organelles that not only help in the breakdown of various substances rather they are the major players in various nervous system pathologies and aging via cathepsins. Hence, targeting these cathepsins may promise better future therapies to improve not only the life span but also the healthspan.
Conflict of Interest Statement
None declared.
Acknowledgements
The authors are thankful to the Vice chancellor of University of the Punjab, Lahore, Pakistan for providing support for the accomplishment of this study.
Authors Contributions
MBK, NF, MHA, RM, ZA and ZS, AA, TS collected the data and performed literature review. MK, NF, MHA and RM write the draft and provided professional guidelines. MBK and NS conceived the idea, approved the final manuscript and supervised the whole project.
References
- Settembre C, Fraldi A, Medina DL, et al. (2013) Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14: 283-296.
- Braun RJ, Sommer C, Leibiger C, et al. (2015) Modeling non-hereditary mechanisms of alzheimer disease during apoptosis in yeast. Microb Cell 2: 136-138.
- Braun RJ, Sommer C, Leibiger C, et al. (2015) Accumulation of basic amino acids at mitochondria dictates the cytotoxicity of aberrant ubiquitin. Cell Rep 10: 1557-1571.
- De Duve C, Pressman B, Gianetto R, et al. (1955) Tissue fractionation studies.6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J 60: 604-617.
- Turk B, Turk V (2009) Lysosomes as "suicide bags" in cell death: myth or reality? J Biol Chem 284: 21783-21787.
- Gasteiger E, Gattiker A, Hoogland C, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784-3788.
- Adams-Cioaba MA, Krupa JC, Xu C, et al. (2011) Structural basis for the recognition and cleavage of histone H3 by cathepsin L. Nat Commun 2: 197.
- Nakanishi H (2020) Microglial cathepsin B as a key driver of inflammatory brain diseases and brain aging. Neural Regen Res 15: 25-29.
- Moore MN (2020) Lysosomes, Autophagy, and hormesis in cell physiology, pathology, and age-related disease. Dose Response 18: 1559325820934227.
- Turk V, Stoka V, Vasiljeva O, et al. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 1824: 68-88.
- Mortimore GE, Reeta Poso A, Lardeux BR (1989) Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev 5: 49-70.
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895-899.
- Cuervo AM (2004) Autophagy: Many paths to the same end. Mol cell biochem 263: 55-72.
- Klionsky DJ (2005) The molecular machinery of autophagy: Unanswered questions. J cell sci 118: 7-18.
- Ciechanover A (2012) Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. BBA-Proteins and Proteomics 1824: 3-13.
- Orenstein SJ, Cuervo AM (2010) Changes in lysosomes and their autophagic function in aging: The comparative biology of lysosomal function. In The Comparative Biology of Aging, Springer 201-226.
- Lopez-Otin C, Blasco MA, Partridge L, et al. (2013) The hallmarks of aging. Cell 153: 1194-1217.
- Cuervo AM, Dice JF (2000) When lysosomes get old. Exp gerontol 35: 119-131.
- Nixon RA, Cataldo AM, Mathews PM (2000) The endosomal-lysosomal system of neurons in Alzheimer's disease pathogenesis: A review. Neurochem res 25: 1161-1172.
- Ivy GO, Schottler F, Wenzel F, et al. (1984) Inhibitors of lysosomal enzymes: Accumulation of lipofuscin-like dense bodies in the brain. Science 226: 985-987.
- Nunomura A, Miyagishi T (1993) Ultrastructural observations on neuronal lipofuscin (age pigment) and dense bodies induced by a proteinase inhibitor, leupeptin, in rat hippocampus. Acta Neuropathol 86: 319-328.
- Hook V, Yoon M, Mosier C, et al. (2020) Cathepsin B in neurodegeneration of Alzheimer's disease, traumatic brain injury, and related brain disorders. Biochim Biophys Acta Proteins Proteom 1868: 140428.
- Meng J, Liu Y, Xie Z, et al. (2020) Nucleus distribution of cathepsin B in senescent microglia promotes brain aging through degradation of sirtuins. Neurobiol Aging 96: 255-266.
- Nakanishi H, Tominaga K, Amano T, et al. (1994) Age-related changes in activities and localizations of cathepsins D, E, B, and L in the rat brain tissues. Exp Neurol 126: 119-128.
- Bednarski E, Ribak CE, Lynch G (1997) Suppression of cathepsins B and L causes a proliferation of lysosomes and the formation of meganeurites in hippocampus. J Neurosci 17: 4006-4021.
- Porta E, Llesuy S, Monserrat AJ, et al. (1995) Changes in cathepsin B and lipofuscin during development and aging in rat brain and heart. Gerontology 41: 81-93.
- Bernstein HG, Kirschke H, Wiederanders B, et al. (1990) Antigenic expression of cathepsin B in aged human brain. Brain res bull 24: 543-549.
- Wang JL, Xu CJ (2020) Astrocytes autophagy in aging and neurodegenerative disorders. Biomed Pharmacother 122: 109691.
- Nakanishi H, Wu Z (2009) Microglia-aging: Roles of microglial lysosome-and mitochondria-derived reactive oxygen species in brain aging. Behav Brain Res 201: 1-7.
- Von Bernhardi R, Eugen-von Bernhardi L, Eugen J (2015) Microglial cell dysregulation in brain aging and neurodegeneration. Front Aging Neurosci 7: 124.
- Halle A, Hornung V, Petzold GC, et al. (2008) The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat immunol 9: 857-865.
- Wendt W, Zhu XR, Lubbert H, et al. (2007) Differential expression of cathepsin X in aging and pathological central nervous system of mice. Exp neurol 204: 525-540.
- Hayashi Y, Koyanagi S, Kusunose N, et al. (2013) The intrinsic microglial molecular clock controls synaptic strength via the circadian expression of cathepsin S. Scientific Reports 3: 2744.
- Hafner A, Glavan G, Obermajer N, et al. (2013) Neuroprotective role of gamma-enolase in microglia in a mouse model of Alzheimer's disease is regulated by cathepsin X. Aging cell 12: 604-614.
- Banay-Schwartz M, DeGuzman T, Kenessey A, et al. (1992) The distribution of cathepsin D activity in adult and aging human brain regions. J neurochem 58: 2207-2211.
- Dorn A, Muller A, Kirschke H, et al. (1986) Immunohistochemical analysis of the distribution of cathepsin D throughout human nervous system with reference to developmental aspects. Biomed biochim acta 45: 1457-1460.
- Kenessey A, Banay-Schwartz M, De Guzman T, et al. (1989) Increase in cathepsin D activity in rat brain in aging. J neurosci res 23: 454-456.
- Matus A, Green GD (1987) Age-related increase in a cathepsin D like protease that degrades brain microtubule-associated proteins. Biochemistry 26: 8083-8086.
- Nakamura YU, Takeda M, Suzuki H, et al. (1989) Age-dependent change in activities of lysosomal enzymes in rat brain. Mechanisms of Ageing and Development 50: 215-225.
- Snyder DS, Whitaker JN (1983) Postnatal changes in cathepsin D in rat neural tissue. Journal of Neurochemistry 40: 1161-1170.
- Haas U, Sparks DL (1996) Cortical cathepsin D activity and immunolocalization in Alzheimer disease, critical coronary artery disease, and aging. Mol Chem Neuropathol 29: 1-14.
- Snyder DS, Simonis S, Uzman BG, et al. (1985) Rat neural tissue cathepsin D: Ultrastructural immunocytochemistry. J Neurocytol 14: 579-596.
- Bi X, Yong AP, Zhou J, et al. (2000) Regionally selective changes in brain lysosomes occur in the transition from young adulthood to middle age in rats. Neuroscience 97: 395-404.
- Jung H, Lee EY, Lee SI (1999) Age-related changes in ultrastructural features of cathepsin B- and D-containing neurons in rat cerebral cortex. Brain Res 844: 43-54.
- Sato Y, Suzuki Y, Ito E, et al. (2006) Identification and characterization of an increased glycoprotein in aging: Age-associated translocation of cathepsin D. Mech Ageing Dev 127: 771-778.
- Benuck M, Banay-Schwartz M, DeGuzman T, et al. (1996) Changes in brain protease activity in aging. J Neurochem 67: 2019-2029.
- Platt FM, Boland B, van der Spoel AC (2012) The cell biology of disease: Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. J Cell Biol 199: 723-734.
- Beltroy EP, Richardson JA, Horton JD, et al. (2005) Cholesterol accumulation and liver cell death in mice with Niemann-Pick type C disease. Hepatology 42: 886-893.
- Hara T, Nakamura K, Matsui M, et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885-889.
- Komatsu M, Waguri S, Chiba T, et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880-884.
- Mizushima N, Levine B (2020) Autophagy in human diseases. N Engl J Med 383: 1564-1576.
- Menezes R, Tenreiro S, Macedo D, et al. (2015) From the baker to the bedside: Yeast models of Parkinson's disease. Microb Cell 2: 262-279.
- Westbroek W, Gustafson AM, Sidransky E (2011) Exploring the link between glucocerebrosidase mutations and parkinsonism. Trends Mol Med 17: 485-493.
- Jung T, Bader N, Grune T (2007) Lipofuscin: Formation, distribution, and metabolic consequences. Ann N Y Acad Sci 1119: 97-111.
- Hohn A, Jung T, Grimm S, et al. (2011) Lipofuscin inhibits the proteasome by binding to surface motifs. Free Radic Biol Med 50: 585-591.
- Madeo F, Zimmermann A, Maiuri MC, et al. (2015) Essential role for autophagy in life span extension. J Clin Invest 125: 85-93.
- Folick A, Oakley HD, Yu Y, et al. (2015) Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347: 83-86.
- Plotegher N, Duchen MR (2017) Mitochondrial dysfunction and neurodegeneration in lysosomal storage disorders. Trends Mol Med 23: 116-134.
- Hughes AL, Gottschling DE (2012) An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492: 261-265.
- Klang A, Schmidt P, Kneissl S, et al. (2014) IgG and complement deposition and neuronal loss in cats and humans with epilepsy and voltage-gated potassium channel complex antibodies. J Neuropathol Exp Neurol 73: 403-413.
- Medina DL, Di PS, Peluso I, et al. (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288-299.
- Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477-513.
- Walter P, Ron D (2011) The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081-1086.
- Terman A, Kurz T, Navratil M, et al. (2010) Mitochondrial turnover and aging of long-lived postmitotic cells: The mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal 12: 503-535.
- Löw P (2011) The role of ubiquitin-proteasome system in ageing. Gen Comp Endocrinol 172: 39-43.
- Kurz T, Terman A, Gustafsson B, et al. (2008) Lysosomes and oxidative stress in aging and apoptosis. Biochim Biophys Acta 1780: 1291-1303.
- Cuervo AM, Dice JF (1998) How do intracellular proteolytic systems change with age? Front Biosci 3: d25-d43.
- Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584: 1393-1398.
- Singh R, Cuervo AM (2011) Autophagy in the cellular energetic balance. Cell Metab 13: 495-504.
- Cuervo AM (2004) Autophagy: In sickness and in health. Trends Cell Biol 14: 70-77.
- Luo L, Qin ZH ( 2019) Autophagy, Aging, and Longevity. Adv Exp Med Biol 1206: 509-525.
- Li WW, Li J, Bao JK (2012) Microautophagy: Lesser-known self-eating. Cell Mol Life Sci 69: 1125-1136.
- Al-Bari MAA, Xu P (2020) Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann N Y Acad Sci 1467: 3-20.
- Xie Z, Klionsky DJ (2007) Autophagosome formation: Core machinery and adaptations. Nat Cell Biol 9: 1102-1109.
- Ohsumi Y (2014) Historical landmarks of autophagy research. Cell Res 24: 9-23.
- Puleston DJ, Simon AK (2015) New roles for autophagy and spermidine in T cells. Microb Cell 2: 91-93.
- Rajawat YS, Hilioti Z, Bossis I (2009) Aging: Central role for autophagy and the lysosomal degradative system. Ageing Res Rev 8: 199-213.
- de CR, Carmona-Gutierrez D, Bernier M, et al. (2014) The search for antiaging interventions: From elixirs to fasting regimens. Cell 157: 1515-1526.
- Tóth ML, Sigmond T, Borsos E, et al. (2008) Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4: 330-338.
- Simonsen A, Cumming RC, Brech A, et al. (2008) Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4: 176-184.
- Singh R, Kaushik S, Wang Y, et al. (2009) Autophagy regulates lipid metabolism. Nature 458: 1131-1135.
- Nezis IP, Stenmark H (2012) p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid Redox Signal 17: 786-793.
- Füllgrabe J, Klionsky DJ, Joseph B (2013) Histone post-translational modifications regulate autophagy flux and outcome. Autophagy 9: 1621-1623.
- Rea SL, Majcher V, Searle MS, et al. (2014) SQSTM1 mutations--bridging Paget disease of bone and ALS/FTLD. Exp Cell Res 325: 27-37.
- Martinez-Vicente M, Talloczy Z, et al. (2010) Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 13: 567-576.
- Geisler S, Holmström KM, Skujat D, et al. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119-131.
- Guo F, Liu X, Cai H, et al. (2018) Autophagy in neurodegenerative diseases: Pathogenesis and therapy. Brain Pathol 28: 3-13.
- Carmona-Gutiérrez D, Bauer MA, Ring J, et al. (2011) The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis 2: e161.
- Lapierre LR, Gelino S, Meléndez A, et al. (2011) Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 21: 1507-1514.
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, et al. (2013) The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 4: 2267.
- O'Rourke EJ, Ruvkun G (2013) MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 15: 668-676.
- Babcock DT, Shen W, Ganetzky B (2015) A neuroprotective function of NSF1 sustains autophagy and lysosomal trafficking in Drosophila. Genetics 199: 511-522.
- Ren J, Yang L, Zhu L, et al. (2017) Akt2 ablation prolongs life span and improves myocardial contractile function with adaptive cardiac remodeling: Role of Sirt1-mediated autophagy regulation. Aging Cell 16: 976-987.
- Ren J, Zhang Y (2018) Targeting Autophagy in Aging and Aging-Related Cardiovascular Diseases. Trends Pharmacol Sci 39: 1064-1076.
- Pucciarelli S, Moreschini B, Micozzi D, et al. (2012) Spermidine and spermine are enriched in whole blood of nona/centenarians. Rejuvenation Res 15: 590-595.
- Eisenberg T, Knauer H, Schauer A, et al. (2009) Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11: 1305-1314.
- Morselli E, Mariño G, Bennetzen MV, et al. (2011) Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol 192: 615-629.
- Baur JA, Sinclair DA (2006) Therapeutic potential of resveratrol: The in vivo evidence. Nat Rev Drug Discov 5: 493-506.
- Lee IH, Cao L, Mostoslavsky R, et al. (2008) A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 105: 3374-3379.
- Morselli E, Maiuri MC, Markaki M, et al. (2010) The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy 6: 186-188.
- Lamming DW, Ye L, Sabatini DM, et al. (2013) Rapalogs and mTOR inhibitors as anti-aging therapeutics. J Clin Invest 123: 980-989.
- Li J, Kim SG, Blenis J (2014) Rapamycin: One drug, many effects. Cell Metab 19: 373-379.
- Mackeh R, Lorin S, Ratier A, et al. (2014) Reactive oxygen species, AMP-activated protein kinase, and the transcription cofactor p300 regulate a-tubulin acetyltransferase-1 (aTAT-1/MEC-17)-dependent microtubule hyperacetylation during cell stress. J Biol Chem 289: 11816-11828.
- Dowling RJ, Zakikhani M, Fantus IG, et al. (2007) Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res 67: 10804-10812.
- Ben SI, Regazzetti C, Robert G, et al. (2011) Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res 71: 4366-4372.
- Zoncu R, Bar-Peled L, Efeyan A, et al. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334: 678-683.
- Mercken EM, Carboneau BA, Krzysik-Walker SM, et al. (2012) Of mice and men: The benefits of caloric restriction, exercise, and mimetics. Ageing Res Rev 11: 390-398.
- Warburton DE, Nicol CW, Bredin SS (2006) Health benefits of physical activity: The evidence. CMAJ 174: 801-809.
- Tam BT, Siu PM (2014) Autophagic cellular responses to physical exercise in skeletal muscle. Sports Med 44: 625-640.
- Hansen M, Rubinsztein DC, Walker DW (2018) Autophagy as a promoter of longevity: Insights from model organisms. Nat Rev Mol Cell Biol 19: 579-593.
- Zhang Q, Wu X, Chen P, et al. (2018) The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent wnt signaling. Cell 174: 870-883.
- Cadwell K, Debnath J (2018) Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol 217: 813-822.
- Cotzomi-Ortega I, Aguilar-Alonso P, Reyes-Leyva J, et al. (2018) Autophagy and its role in protein secretion: implications for cancer therapy. Mediators Inflamm 2018: 4231591.
- Hood S, Amir S (2017) The aging clock: Circadian rhythms and later life. J Clin Invest 127: 437-446.
- Panwar P, Hedtke T, Heinz A, et al. (2020) Expression of elastolytic cathepsins in human skin and their involvement in age-dependent elastin degradation. Biochim Biophys Acta Gen Subj 1864: 129544.
- Yue X, Jiang H, Xu Y, et al. (2020) Cathepsin K deficiency impaired ischemia-induced neovascularization in aged mice. Stem Cells Int 2020: 6938620.
- Hu WX, Li H, Jiang JZ (2020) MiR-491-3p is down-regulated in postmenopausal osteoporosis and affects growth, differentiation and apoptosis of hFOB1.19 cells through targeting CTSS. Folia Histochem Cytobiol 58: 9-16.
- Fan K, Wu X, Fan B, et al. (2012) Up-regulation of microglial cathepsin C expression and activity in lipopolysaccharide-induced neuroinflammation. J Neuroinflammation 9: 96.
- Leeman DS, Hebestreit K, Ruetz T, et al. (2018) Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359: 1277-1283.
Corresponding Author
Muhammad Babar Khawar, State Key Laboratory of Stem cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, P.R. China; University of Chinese Academy of Sciences, Beijing 100049, P.R. China
Muddasir Hassan Abbasi, Department of Zoology, University of Okara, Okara, Pakistan;
Nadeem Sheikh, Cell and Molecular Biology Research Laboratory, Institute of Zoology, University of the Punjab, Lahore, Pakistan
Copyright
© 2021 Khawar MB. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.