The Association between IMPDH II Gene 3757t > C and Ugt1a9 Gene T-275a Polymorphisms with Plasma Mycophenolic Acid among Renal Transplant Recipients
Abstract
Introduction
ESRD is a major health problem imposing great burden on the society. Dialysis either haemodialysis or peritoneal and transplantation are the two alternating treatment modalities. Transplantation became the preferred solution with lifetime monitoring immunosuppressive drugs. Urinary CXCL10 is used for early detection of renal rejection. Mycophenolate is one of the important immunosuppressive drugs used among renal transplant recipients. IMPDH II T3757C and UGT1A9 T275A are important SNPs responsible for MPA pharmacokinetic and inter-individual response variability. The study aimed to evaluate the association between T3757C and T275A SNPs and level of MPA among 50 Egyptian renal transplant recipients from Alexandria University Hospital.
Methods
Real Time-PCR was performed to detect IMPDH II (rs11706052T > C) and UGT1A9 (rs6714486T > A) SNPs, MPA measured by HPLC and Urinary CXCL10 level evaluated by ELISA.
Findings
Variant genotypes in both SNPs IMPDH T3757C and UGT1A9 T275A (CC and AA, respectively) were statistically significantly associated with kidney rejection and lower values of MPA AUC. Linear regression analysis was performed, and it was found that the strongest predictive parameters for lower MPA AUC were the UGT1A9 T275A SNP followed by CXCL10 then IMPDH II T3757C and the lowest predictive parameter was serum creatinine. Urinary CXCL-10 value of 106.58 pg/ml was found to early predict the kidney rejection with a high sensitivity and specificity.
Discussion
Renal transplant recipients should be genotyped before taking MPA to prevent renal allograft rejection. Graft could be monitored by urinary level of CXCL10 with high specificity and sensitivity.
Keywords
End stage renal disease (ESRD), Inosine monophosphate dehydrogenase ii (IMPDH II), Uridine diphosphate glucuronosyl transferases (UGT1A9), Single nucleotide polymorphisms (SNPs), Mycophenolate (MPA), Real time polymerase chain reaction (RT-PCR), High pressure liquid chromatography (HPLC), Enzyme linked immunosorbent assay (ELISA)
Introduction
Chronic kidney disease is a worldwide problem that leads to end-stage renal disease (ESRD). ESRD is considered the 9th cause of death. Also, it is ranking 17th among causes of disability [1].
Dialysis and transplantation are the therapeutic modalities for ESRD. However, dialysis either hemodialysis or peritoneal may expose the patients to many hazards. Nowadays, transplantation has become the treatment of choice for ESRD [2].
The cornerstone of successful transplantation is the introduction of the accurate dose of immunosuppressive drugs at the appropriate time. The use of immunosuppressive drugs should be monitored periodically due to their narrow safety margin. Therapeutic drug monitoring is essential to avoid overdosing toxicity or under-dosing rejection [3].
The chemokine CXCL10 also known as interferon-inducible protein 10 is a promising non-invasive urinary marker for acute rejection. It may indicate the main candidates for the biopsy procedure which is the gold standard tool for defining the acute rejection [4].
The protocol regimen widely used is the combination of tacrolimus and mycophenolate mofetil (MMF) or mycophenolate soduim. They have largely replaced cyclosporine and azathioprine together with prednisone which is considered the most prescribed treatment in developed countries [5].
MMF is metabolized to its active form the mycophenolic acid (MPA). MPA has a selective, reversible inhibitory effect on the inosine monophosohate dehydrogenase (IMPDH II). This enzyme together with the uridine Glucuronosyltransferase (UGT1A9) are two key enzymes important for the drug action. IMPHD II is responsible for the proliferation of the T and B lymphocyte through the de novo pathway for the generation of guanosine. When IMPDH II is inhibited by MPA, this results in arrest of the immune cells. UGT1A9 is the metabolizing enzyme responsible for the clearance of the drug through the glucuronidation of the active compound into its inactive 7‐O‐glucuronide [6].
Monitoring of MPA by high pressure liquid chromatography (HPLC) and enzyme-multiplied immunoassay technique is important to minimize the side effects such as anemia, leucopenia, gastrointestinal upsets and urinary tract infection [7].
Several non-genetic factors may lower the MPA level such as GIT disturbances and drug interactions. Many single nucleotide polymorphisms (SNPs) have been found in IMPDH II and UGT1A9 genes controlling the drug action and metabolism [8].
IMPDH II T3575C leads to increase in the IMPDH II enzyme activity and thus patients carrying this SNP should have higher dose of the drug to fulfil the complete immune response suppression and prevent the rejection [9].
UGT1A9 T275A increases the clearance of the MPA thus lowering its dose in the blood of the patients. This may predispose those patients expressing the mutant genotype to rejection [10].
Aim of the Study
Our study aimed to evaluate the association between T3757C and T275A polymorphisms of the IMPDH II and UGT1A9 genes, respectively and level of MPA among a sample of renal transplant Egyptian recipients.
Subjects
This study was carried out on 50 Egyptian patients aged above 18 years who underwent kidney transplantation. All patients received full dose of the mycophenolate for at least 3 months post-transplantation. Pregnant women, simultaneous transplantation of any other organ, those not receiving MPA and patients with GIT problems have been excluded from the study. They were recruited from the Outpatient Clinic of Renal Transplantation Unit in Alexandria University Hospital. The study was approved by the Ethics Committee of Alexandria University, Egypt. Informed consent was obtained from all individual participants included in the study.
Methods
All patients were subjected to the following:
- Full history.
- Thorough clinical examination.
- Routine laboratory investigations were taken from the records of patients.
- Genetic study.
DNA extraction
Genomic DNA was extracted from EDTA whole blood samples using QIAamp DNA blood mini Kit 50 (Qiagen, CA, USA). The concentration and purify of extracted DNA was assessed using a Nanodrop 2000 spectrophotometer and then stored at -20 ℃ till genotyping.
Genotyping
The following polymorphisms were detected by 5' nuclease allele discrimination assay using real-time PCR; IMPDH II (rs11706052T > C) SNP and UGT1A9 (rs6714486T > A) SNP. Rotorgene Q Real time PCR system was employed to obtain specific sequences amplification according to the following steps: 10 µl of TaqMan® universal PCR Master Mix (20X) was added followed by 0.5 µl of specific assay mix (40X) (T275A, T3757TC) containing primers and probes. Then 1-20 ng of extracted DNA and sterile water were added to complete a total volume of 20 µl. The thermal profile was as such: An initial step at 95 ℃ for 10 minutes (hold), followed by 40 cycles of denaturing to 95 ℃ for 15 seconds and annealing/extension at 60 ℃ for 1 minute.
- Measurement of the level of MPA was assessed by HPLC using ClinRep® HPLC Complete Kit for Mycophenolic Acid in plasma from RECIPE®.
- Measurement of the level of CXCL10 in urine using Quantikine® ELISA, Human CXCL10/IP-10 Immunoassay, was done.
Statistical analysis of the data
Collected data and results were analyzed using IBM SPSS software package version 20.0 (Armonk, NY: IBM Corp). Qualitative data were described using number and percent. The Kolmogorov-Smirnov test was used to verify the normality of distribution. Quantitative data were described using range, mean, standard deviation and median. Significance of the obtained results was judged at the 5% level.
Results
Demographic data and clinical data
60% of the studied population were males while 40% were females with mean age 39.72 ± 10.40 years. Patients received: MPA with either cyclosporine or tacrolimus plus prednisone. Mycophenolate Area Under Curve (MPA AUC) ranged 23.87-1872.04 (× 103) mV/min with mean of 519.52 ± 502.48 (× 103) mV/min. 10% of patients experienced biopsy-proven acute rejection. The CXCL10 urinary level ranged 0.15-169.05 (pg/ml) with mean of 52.79 ± 40.92 pg/ml (Table 1).
Genetic study
As regards to IMPDH II T3757C, 66% were homozygous wild (TT), 28% were heterozygous (TC) and 6% were homozygous variant (CC). As for UGT1A9 T275A SNP, 50% were homozygous wild (TT), 46% were heterozygous (TA) and 4% were homozygous variant (AA). Both IMPDH II T3757C and UGT1A9 T275A were in Hardy-Weinberg equilibrium (p = 0.377 and 0.238, respectively).
Relation between clinical outcome and various studied parameters
The kidney rejection was significantly associated with variant genotypes CC and AA in both IMPDH II T3757C and UGT1A9 T275A (p < 0.001 and p = 0.001, respectively).
Also, MPA AUC showed a statistical significant lower level among patients with graft failure with a mean of 45.7 ± 15.3 (p < 0.001) as shown in Table 2.
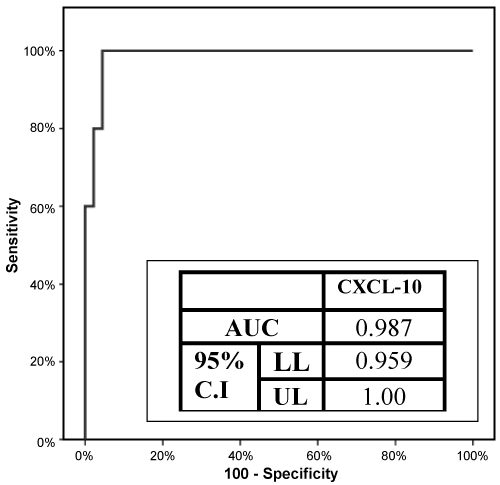
Urinary levels of CXCL10 were significantly higher in patients with allograft failure (132.70 ± 24.27 pg/ml) compared to those with normal kidney function (43.91 ± 31.70 pg/ml) at p < 0.001. ROC curve for CXCL10 was done to predict kidney rejection at the cut-off value of 106.58 pg/ml with highest sensitivity (80%), specificity (95.56%) and AUC was 0.987 (95% CI 0.959-1.00) demonstrated in Figure 1.
Relation between MPA AUC and various studied parameters
Patients expressing variant genotypes in both SNPs; IMPDH II T3757C and UGT1A9 T275A had lower MPA AUC with mean 42.46 (× 103) ± 17.07 (× 103) and 50.52 (× 103) ± 16.45 (× 103), respectively compared with mean of patients expressing wild genotypes 711.10 (× 103) ± 514.43 (× 103) and 874.56 (× 103) ± 483.82 (× 103), respectively (p < 0.001).
We found a significant negative correlation between the CXCL10 and MPA AUC (p < 0.001). Multivariate linear regression was done considering (IMPDH II T3757C and UGT1A9 T275A) as well as urea, creatinine, eGFR and CXCL10 as confounding factors while MPA AUC as dependent variable. UGT1A9 T275A was the strongest predictive parameters for lower MPA AUC followed by CXCL10 then IMPDH II T3757C and lowest one was creatinine (t = 6.562, 3.786, 3.757 and 3.725, p < 0.001, < 0.001, 0.001 and 0.001, respectively) (Table 3).
Discussion
ESRD is a major health burden on the society. Transplantation became a mean to improve patients' quality of life. Therapeutic drug monitoring and personalized medicine have been considered main targets in renal transplantation success. Several biomarkers have been described to predict the renal allograft rejection through gene expression profiling, assessment of regulatory T cells and the urinary chemokine CXCL10 [1,6].
Mycophenolate mofetil and mycophenolate sodium are cornerstones in the protocol of treatment of renal transplant. Inter-individual variability of pharmacokinetics of the drug influences its efficacy and necessitates good monitoring and highlighting factors influencing MPA level [6].
Non-genetic factors may interfere with the measurement of the drug such as hypoalbuminemia, hyperbilirubinemia and drug interactions. Also, IMPDH II and UGT1A9 genes are responsible for two key enzymes in the metabolism of the drug [7].
Regarding demographic data in our study
The homozygous wild genotype in both SNPs (TT) had the highest frequency followed by the heterozygous (TC and TA) and small percent were homozygous variant (CC and AA) among IMPDH II and UGT1A9, respectively. Both SNPs did not show any significant deviation from Hardy-Weinberg equilibrium.
In agreement with our study Grinyo J, et al. [11] studied IMPDH II T3757C, detected only 1% of CC genotype so they were pooled with the heterozygous TC representing 28 patients. The remaining patients 193 found to express the TT genotype. No deviation found from Hardy-Weinberg equilibrium.
Van Schaik R, et al. [12] supported our study when evaluated UGT1A9 T275A, they showed that the highest frequency was the homozygous wild type (TT), followed by the heterozygous (TA) and no homozygous variant was found.
Studying the UGT1A9 T275A Kuypers DR, et al. [13] agreed with us and found 79 patients with homozygous wild (TT), 15 patients heterozygous (TA) and homozygous variant (AA) was 1 patient and they were in Hardy-Weinberg equilibrium.
Regarding the relation between the clinical outcome and various studied parameters
We found that kidney rejection was significantly associated with the homozygous variant (CC and AA) and the heterozygous genotypes (TC and TA) (p < 0.001 and p = 0.001) of both SNPs; IMPDH II T3757C and UGT1A9 T275A, respectively. This was explained by that IMPDH II T3757C variant (CC) reduced the drug efficacy by increasing the IMPDH II enzyme activity and this lead to acute rejection. On the other hand the homozygous variant (AA) of the UGT1A9 T275A increased the clearance of the drug by glucuronidation which also lead to kidney rejection. Patients with graft rejection were significantly associated with lower value of MPA AUC (p < 0.001). CXCL10 was significantly higher among patients with kidney rejection (p < 0.001). ROC curve which is a fundamental tool for assessing the performance of CXCL10 in prognosis and diagnosis of kidney rejection was done and identified 106.58 pg/ml as a cut-off value of urinary CXCL10 to predict kidney rejection with sensitivity of 80% and with specificity of 95.56% with AUC value of 0.987 (CI: 95%, p < 0.001).
In concordance with our study Ciliao HL, et al. [14] evaluated different SNPs among genes related to the transplantation in 246 transplanted patients (35% diagnosed with graft rejection). It was concluded that UGT1A9 T275A (TA and TT) were associated with kidney rejection. This was attributed to increase in the glucuronidation of MPA by the enhanced expression and activity of UGT1A9 enzyme which leads to reduced concentration of MPA and therefore diminished its immunosuppressive activity, promoting the initiation of organ rejection episodes.
Similarly, Raza A, et al. [15] investigated the relation of urinary CXCL10 with the clinical outcome of rejection and found a statistical significant difference in urinary CXCL10 levels between the allograft rejection and control cases. They had a cut-off value of 27.5 pg/ml of urinary CXCL10 with highest sensitivity of 72% and specificity of 71% with AUC value of 0.74 ± 0.04 at confidence interval of 95%.
Moreover, Rabant M, et al. [16] concluded that increased urinary CXCL10 may be associated with any type of alloimmune injury. According to them, a biopsy must be performed in cases of acute allograft dysfunction to assess the mechanism of injury and the proper therapeutic interventions.
In agreement with us, Mao Y, et al. [17] stated that urinary CXCL10 levels increases with acute renal allograft rejection. They also confirmed the promising role of the chemokine CXCL10 in monitoring the graft rejection.
In addition, Jackson JA, et al. [18] found that urinary CXCL10 was markedly elevated in urine samples among the studied population suffering from acute rejection.
Also, Oetting WS, et al. [19] proved that a weak association was found between IMPDH II T3757C and acute rejection. This was explained by reporting that the presence of the variant allele C either in the heterozygous TC genotype or the homozygous variant CC result in a reduced response to MPA compared to homozygous wild TT resulting in increased risk of acute rejection.
When genotyping the DNA of 20 volunteers for IMPDH II T3757C, Winnicki W, et al. [10] agreed with our study that patients carrying the variant genotype had increase enzyme activity and reduced efficacy of MPA drug. They concluded that heterozygous TC genotype and homozygous variant CC genotype must have higher doses of MPA drug than homozygous wild TT.
In concordance with our study, Grinyo J, et al. [11] analysed IMPDH II T3757C in 237 patients and detected those patients with TC or CC were more likely to end by biopsy proved acute rejection (BPAR) than those with homozygous wild TT. They proved a statistical significant association with BPAR post-transplant.
In contrast to us, a study conducted on a large group of renal transplant recipients by Gensburger O, et al. [20] found no association between IMPDH II T3757C and BPAR. This was attributed to different ethnicity of the studied populations and to a limited number of populations studied by the others who found association.
As regards to the relation between the MPA AUC and various studied parameters
We found that MPA AUC was significantly lower among patients expressing the homozygous variant (CC and AA) for both studied SNPs; IMPDH II T3757C and UGT1A9 T275A, (p < 0.001) with mean values 42.46 ± 17.07 and 50.52 ± 16.45, respectively. We found that urinary CXCL10 was significantly associated with lower level of MPA AUC (p < 0.001). Finally, multivariate linear regression analysis proved that the strongest statistically significant predictive parameters for lower MPA AUC was UGT1A9 T275A followed by CXCL10 then IMPDH II T3757C and the lowest predictive parameter was creatinine.
This was supported by Kuypers DR, et al. [13] who showed a significant decrease in MPA level in those who carried the variant allele compared with those homozygous wild type (TT). This was explained by the significantly increase of the estimated total body clearance in recipients with the UGT1A9 T275A. Also, they performed multiple regression analysis to assess the role of UGT1A9 T275A promoter region SNP in inter-individual variability of MPA pharmacokinetics with other clinical covariates. They stated that UGT1A9 T275A was the most significant independent predictor for MPA AUC.
Similarly, Van Schaik R, et al. [12] found a lower MPA AUC early post-transplantation and higher rejection rate among patients carrying the heterozygous UGT1A9 T275A (TA) when compared with those homozygous wild (TT). However, they did not found any statistical significant difference.
Similarly, Ruiz J, et al. [21] stated that patients who are wild (TT) for UGT1A9 T275A had higher level of MPA, than those with heterozygous genotype (TA) although statistical significance was not reached.
In agreement to our study, Li P, et al. [22] detected higher clearance in UGT1A9 275T >A carriers that lead to a decrease in MPA-AUC up to 50%.
In concordance to our study, Mazidi T, et al. [23] concluded that patients carrying UGT1A9 T275A who were heterozygous (TA) had significantly lower AUC for MPA in comparison to the homozygous wild (TT). This was attributed to increase in clearance of the drug and increased glucuronidation compared to wild type patients.
Sanchez-Fructuoso AI, et al. [24] found that the T-275A mutation in UGT1A9 showed a lower MPA exposure level which leads do decrease in the efficacy of the immunosuppressive drug and patients were more liable to graft failure.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflicts of interest that are directly relevant to the content of this article.
Ethical standards
This research was conducted on human participants. The study was approved by the Ethics Committee of Alexandria University, Egypt.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
References
- Stanifer JW, Muiru A, Jafar TH, et al. (2016) Chronic kidney disease in low- and middle-income countries. Nephol Dial Transplant 31: 868-874.
- Yamamoto S, Kazama JJ, Wakamatsu T, et al. (2016) Removal of uremic toxins by renal replacement therapies: a review of current progress and future perspectives. Renal Replacement Ther 2: 43-47.
- Khatri M (2017) Dealing with side effects after an organ transplant.
- Schaub S, Nickerson P, Rush D, et al. (2009) UrinaryCXCL9 and CXCL10 levels correlate with the extent of subclinical tubulitis. Am J Transplant 9: 1347-1353.
- Bestard O, Campistol JM, Morales JM, et al. (2012) Advances in immunosuppression for kidney transplantation: New strategies for preserving kidney function and reducing cardiovascular risk. Nefrologia 32: 374-384.
- (2013) Product Monograph. CellCept®, Hoffmann-La Roche Ltd, 1-59.
- Maddela R, Pilli NR, Maddela S, et al. (2017) A novel and Rapid LC-MS/MS assay for the determination of mycophenolate and mycophenolic acid in human plasma. J Young Pharm 9: 106-113.
- Tornatore KM, Sudchada P, Attwood K, et al. (2013) Race and drug formulation influence on mycophenolic acid pharmacokinetics in stable renal transplant recipients. J Clin Pharmacol 53: 285-293.
- Winnicki W, Weigel G, Sunder-Plassmann G, et al. (2010) An inosine 5'-monophosphate dehydrogenase 2 single-nucleotide polymorphism impairs the effect of mycophenolic acid. Pharmacogenomic J 10: 70-75.
- Xie XC, Wang HY, Li HL, et al. (2015) Associations of UDP glucouronosyltransferases polymorphisms with mycophenolate mofetil pharmacokinetics in Chinese renal transplant patient. Acta Pharmacol Sin 36: 644-650.
- Grinyo J, Vanrenterghem Y, Nashan B, et al. (2008) Association of four DNA polymorphisms with acute rejection after kidney transplantation. Transpl Int 21: 879-891.
- van Schaik R, van Agteren M, de Fijter J, et al. (2009) UGT1A9 -275T>A/-2152C>T polymorphisms correlate with low MPA exposure and acute rejection in MMF/tacrolimus-treated kidney transplant patients. Clin Pharmacol Ther 3: 319-327.
- Kuypers DR, Naesens M, Vermeire S, et al. (2005) The impact of uridine diphosphate- glucouronyltransferase 1A9 (UGT1A9) gene promoter region single nucleotide polymorphisms T-275A and C-2152T on early mycophenolic acid dose-interval exposure in de novo renal allograft recipients. Clin Pharmacol Ther 78: 351-361.
- Ciliao HL, Camargo-Godoy RBO, de souza MF, et al. (2017) Association of UGT2B7, UGT1A9, ABCG2, and IL23R polymorphisms with rejection risk in kidney transplant patients. J Toxicol Environ Health A 80: 661-671.
- Raza A, Firasat S, Khaliq S, et al. (2017) The association of urinary interferon-gamma inducible protein-10 (IP10/ CXCL10) levels with kidney allograft rejection. Inflamm Res 66: 425-432.
- Rabant M, Amrouche L, Lebreton X, et al. (2015) Urinary chemokine C-X-C motif chemokine 10 independently improves the non-invasive diagnosis of antibody mediated kidney allograft rejection. J Am Soc Nephrol 26: 2840-2851.
- Mao Y, Wang M, Zhou Q, et al. (2011) CXCL10 and CXCL13 expression were highly up-regulated in peripheral blood mononuclear cells in acute rejection and poor response to anti-rejection therapy. J Clin Immunol 31: 414-418.
- Jackson JA, Kim EJ, Begley B, et al. (2011) Urinary chemokines CXCL9 and CXCL10 are non invasive markers of renal allograft rejection and BK viral infection. Am J Transplant 11: 2228-2234.
- Oetting WS, Schladt DP, Leduc RE, et al. (2011) Validation of single nucleotide polymorphisms (SNPs) associated with acute rejection in kidney transplant recipients using a large multi-center cohort. Transpl Int 12: 1231-1238.
- Gensburger O, Van Schaik RN, Picard N, et al. (2010) Polymorphisms in type I and II inosine monophosphate dehydrogenase genes and association with clinical outcome in patients on mycophenolate mofetil. Pharmoacogenet Genomics 20: 537-543.
- Ruiz J, Herrero MJ, Boso V, et al. (2015) Impact of Single Nucleotide Polymorphisms (SNPs) on immunosuppressive therapy in lung transplantation. Int J Mol Sci 16: 20168-20182.
- Li P, Shuker N, Hesselink DA, et al. (2014) Do Asian renal transplant patients need another mycophenolate mofetil dose compared with Caucasian or African American patients? Steunstichting ESOT 27: 994-1004.
- Mazidi T, Rouini MR, Ghaheremani MH, et al. (2013) Impact of UGT1A9 polymorphism on mycophenolic acid pharmacokinetic parameters in stable renal transplant patient. Iran J Pharm Res 12: 547-556.
- Sanchez-Fructuoso AI, Maestro ML, Calvo N, et al. (2009) The prevalence of uridine diphosphate- glucuronosyltransferase 1A9 (UGT1A9) gene promoter region single nucleotide polymorphisms T-275A and C-2152T and its influence on mycophenolic acid pharmacokinetics in stable renal transplant patients. Transplant Proc 41: 2313-2316.
Corresponding Author
Iman Said Gharraf, Faculty of Medicine, Department of Clinical and Chemical Pathology, University of Alexandria, Egypt.
Copyright
© 2018 Sharaki OA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.