Rice Bran Fermented with Newly Isolated Lactobacillus Plantarm IICF2-1 Influences the Growing Properties and the Intestinal Bacteria in Growing Piglets
Abstract
Objective: The objective of this study was to investigate the influence of fermented rice bran with Lactobacillus plantarum ⅡCF2-1 to the growth performance and changes of intestinal bacteria of piglets.
Methods: Rice bran (RB) fermented with a newly isolated Lactobacillus plantarum (L. plantarum ⅡCF2-1) was prepared, and added to basic feed (5%, w/w), then fed to growing piglets for 40 days. The growing performance and the changes of intestinal bacteria of the piglets were studied.
Results: L. plantarum ⅡCF2-1 showed rapid growth (generation time ≈ 37 min), short lag phase (=1.5 h), low pH (< 4.5) and high L-lactic acid yield ( ≈ 6.5 g/L). The content of acid soluble protein was 78.3% higher and crude fiber was 32.4% lower in fermented rice bran (FRB) than RB. During the 40 days feeding, average daily gain of the piglets were higher (by 4.1 %), and the ratios of feed to gain were lower (by 3.2 %) in FRB group than the basal diet group. The genera number in the fecal of FRB fed piglets at day 40 (308) was greater than that at day 0 (256). Among the total 331 genera, abundances of 202 were increased and 29 were decreased. The abundances of many beneficial bacterium including Clostridium sensu-stricto 1, Helcococcus and Bifidobacterium were increased, of many pathogenetic genera including Enterobacter, Escherichia-Shigella were decreased.
Conclusion: The quality of RB has been improved by fermentation, and the FRB has the potential to improve growing pigs' health.
Keywords
Rice bran, Lactobacillus plantarum, Solid-state fermentation, Intestinal bacteria
Introduction
There are a large number of microorganisms in the intestinal tract of animals. They are in close contact with the cells of the body and have the effects of nutrition, immunity and symbiosis on the host. Lactobacillus plantarm is an important probiotic, which is widely present in dairy products [1,2], vegetables [3], other feed [4] and gastrointestinal tract [5]. The dietary L. plantarum could promote the growing of the pigs by reduce the pigs' odor emissions [6], reduce the Enterobacteriaceae in the intestines [6], promote the increase of pigs' villus height [7], and so on.
Rice bran is one of the major by-products of rice milling process and the most abundant agricultural wastes, which is rich in proteins, oil, dietary fiber and other nutrients [8,9]. However, its application shows some limitations due to the rancidity in a short period of time, a certain amount of insoluble fiber and phytic acid and so on. It may be practicable that rice bran is utilized by applying biotechnological methods such as modern fermentation. Fermentation of rice bran by probiotics such as L. plantarum, Aspergillus oryzae and Rhizoctonia oryzae is a valuable technique for utilization of agro-industrial by-products to produce value added products [10] accompanied with higher yields and productivities of valuable compounds such as enzymes, organic acids and many other bioactive compounds [11], which have antioxidant as well as skin-related enzyme inhibition activities [9,12].
In addition, a number of studies suggested that the fermented feed/food exert some significant influence on the synthesis of some nutrients and digestibility of energy for animals, which were carried out by the effects of intestinal microorganism [13]. For example, rice bran fermented with L. plantarum B2 showed high characteristics to improve intestinal environment and produce short chain fatty acid (acetic, propionic and butyric acids) [14], as well as improve the intestinal environment and blood lipid condition [15].
In this study, a new L. plantarum, named L. plantarum ⅡCF2-1 was isolated from a silage sample. Rice bran was fermented by L. plantarum ⅡCF2-1 and was added to the feed for the growing pigs. The main objective of this work was to understand the influences of the L. plantarum ⅡCF2-1 fermented rice bran to the growth and health parameters of piglets, including changes of microbiotic diversity and quantity of L. plantarum in the intestinal tract. This work would provide new materials for the development and utilization of novel fermented feed for piglets.
Materials and Methods
All the experiment involving animals were approved by the Academy of National Food and Strategic Reserves Administration Committee on the Use of Experimental Animals (Protocol number: LA2019-0106), and conform to the "Laboratory animal control ordinance (2017)" and "Guidelines for ethical review of experimental animal welfare (GB/T 35892-2018)" of China.
Isolation and screening of L. plantarum ⅡCF2-1
L. plantarum ⅡCF2-1, obtained from a silage sample, was cultivated in improved MRS medium for replacing main carbon source (1000 mL water, 10 g rice bran, 10 g peptone, 5 g yeast extract, 5 g sodium acetate, 2 g K2HPO4, 2 g diammonium citrate, 1 mL Tween 80, 0.58 g MgSO4·7H2O, 0.25 g MnSO4·H2O, pH 7.0). After incubated at 35 ℃ for overnight, the characteristics of this strain were investigated, including the growth rate, lag phase, L-lactic acid yield and pH change and so on according to the reference [16].
Solid state fermentation of rice bran
The fermentation of 2 t rice bran (divided into about 7 portions) was carried out on a pilot scale using 500 L solid state fermentation (SSF) bioreactor. In one trial, 300 kg rice bran was mixed with 20 % water (w/w), 10 % L. plantarum ⅡCF2-1 broth (w/w) and 5 ‰ acid protease (w/w; 50 u/mg). The inoculated bran was fermented for 2 days at the relative anaerobic environment. The fermented rice bran was evaluated by detecting the content of crude protein, crude fiber, acid soluble protein and fatty acid value according to Chinese national standards on feed.
Feeding trial
The trial was conducted at the feeding base under standardized conditions. A total of 100 weaned piglets of the similar weight, age and health were selected for use. All the pigs were maintained in a controlled conventional environment. The experimental diets and water were available ad libitum during the entire experimental period. Using a single factor experimental design, piglets were randomly divided into 2 groups with 5 replicates and 10 piglets per replicate:Group A: basal diet as negative control; Group B: 50g fermented rice bran per kg basal diet. The standard diet was formulated to meet the nutrient needs suggested by the NRC (National Research Council,1998; Table S1). Body weights and feed weights were determined every 7 ds. Average daily gain, average daily feed intake, and ratio feed to gain were calculated from d 0 through d 40 using EXCEL software.
The analysis of microbial diversity
20 samples were taken at d 0 and d 40, respectively in Group B and were analyzed. The samples were cooled to 4 ℃ immediately after collection, delivered within 24 h, and frozen at -80 ℃ upon receipt until the analysis. Bacterial DNA of animal fecal samples was extracted using DNA kit (Omega Bio-tek Inc., Norcross, GA, USA) according to the manufacturer's instructions. A total of 467 bp of the V3-V4 region of the bacteria 16S's ribosomal RNA gene were amplified by PCR, using primers 338F 5'-ACTCCTACGGGAGGCAGCAG)-3' and 806R 5'-GGACTACHVGGGTWTCTAAT-3'. The PCR reaction system (20 μL) consisted of 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, 10 ng of template DNA and UV-sterilized water. The PCR products were sent to Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) for sequencing on an Illumina hiSeq platform. The raw fastq files were multiplexed and quality-filtered using QIIME (version 1.17) with the criteria. Operational Units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.1, http://drive5.com/uparse/). Chimerical sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) using a confidence threshold of 70% [17].
Quantitative real-time PCR
Fecal samples were collected, and DNA was extracted using the Fast DNA Spin Kit for Soil (catalog no. 6560-200; MP Biomedical) following the manufacturer's instructions. The region of the 16S rRNA was amplified by PCR from microbial genome DNA using the following primers: forward primer, 5′-TGGATCACCTCCTTTCTAAGGAAT-3′; reverseprimer, 5′-GTTCTCGGTTTCATTATGAAAAAATA-3′. The cycle parameters were modified according to the report of Water, et al. and Haarman [18,19]. PCR products were gel-purified using Gene Clean Turbo (catalog no. 111102400; MP Biomedicals) and quantified with an Quant-iTPicoGreen dsDNA Assay Kit (catalog no. P7589; Life Technologies).The reactions is carried out by ABI 7500 Real time PCR.
Data analysis
Significance tests and Fitting Analysis were performed using SPSS16.0 software and OriginPro software, respectively. Cluster analysis was performed using R software.
Results
Isolation, identification, and properties of L. plantarum ⅡCF2-1
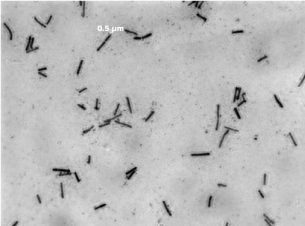
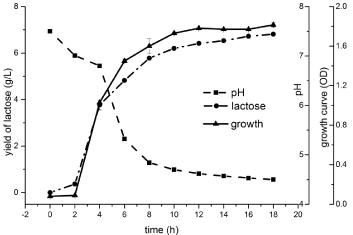
In the process of collecting and analyzing silage samples (the main ingredients of many samples include rice bran), a total of 73 potentially probiotic strains were isolated. 20 strains were identified as L. plantarum by 16S rRNA gene sequencing, and were regarded as safe species and allowed as feed additive in China. Among them, L. plantarum ⅡCF2-1 was preliminarily determined as a new strain by comparing with NCBI database. In the selected medium, L. plantarum ⅡCF2-1 was singly or in pairs , with a rod-shaped of diameter of 0.5 μm and length of 2-5μm (Figure 1). The growth of L. plantarum ⅡCF2-1 fits Gomperdz model [20] as (equation 1) and Figure 2 shows, ODMAX = 1.815 = 4.1×109 cfu/m L. In equation 1, X is the concentration of biomass; A is the maximum biomass; t is the cell culture time (h); λ is the cell growth delay time (h); μm is the maximum specific growth rate (H-1).
The initial pH value was 7.50. It was decreased from 7.50 to 6.80 during 0 ~ 4 h. Then the pH value decreased exponentially due to the large amount of lactic acid produced by the fermentation of strain. From the statistical results of the viable bacteria, it displayed rapid growth (generation time about 37 min), short lag phase (1.5h) and high L-lactic acid yield (up to 6.5 g/L). After 10 h, it entered the stable phase, with the highest number of viable bacteria being 4.1 × 109 cfu/mL.
Chemical and microbiological analysis of the fermented rice bran
Samples of non-fermented and fermented rice bran were tested for nutritional changes by evaluating the content of Lactobacillus and mould, crude protein and acid soluble protein (Table 1 and Figure 3).
The pH value decreased from 7.03 to 4.47 at 2 days' fermentation, which was 2.56 lower than Non-fermented rice bran. And the corresponding viable cell number of L. plantarum ⅡCF2-1 increased significantly (p < 0.05) from 1.12 × 108 cfu/g (corresponding to 8.05 in Table 2) to 6.92 × 108 cfu/g (corresponding to 8.84 in Table 2). In the subsequent 10 days' experiment, the cell numbers of L actobacillus and mould did not change much, only the pH value was continued to decrease and reached a minimum of 4.06 on the 12th day. Therefore, the most suitable fermentation time is 2 days. The fermented rice bran had good smell, homogeneous color and suitable humidity for feed.
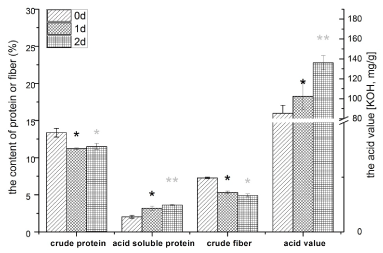
The acid soluble protein and crude fiber, which were the two important indexes for evaluating the quality of fermented feed, were found to be significantly (p < 0.05) changed in all samples in 2 days fermentation as Figure 3 shows. The content of acid soluble protein increased significantly from 2.07% to 3.69 % (p < 0.05), and that of crude fiber decreased significantly from 7.4 % to 5.0% (p < 0.05). Furthermore, the increasing of the fatty acid value and over-oxidation had been inhibited (136.1 mg(KOH)/g), compared to non-fermented counterpart (160.0 mg(KOH)/g, obtained in the pre-experiment). Differ to the result of the former, the fermented rice bran showed the content of crude protein decreased significantly from 13.6% to 11.7 % (p < 0.05) because of the function of proteases.
Growth performance of piglets
The growth performances of the 2 feeding treatment groups showed difference during the whole experiment (Table 2). Over the 40 ds of the feeding, the body weight gain was 17.24 kg (from 10.07 kg to 27.31 kg) in group B (experiment group), which was better than 16.69 kg in group A (control group, 26.70 kg to 10.01 kg). Average daily gain of the piglets in groups B (620.7g ± 20.2g) was significantly higher (P < 0.05) than that in group A (596.1g ± 16.0g). The average daily feed intake in group B (935.5 ± 41.2) was higher than that in group A (931.4 ± 36.0). However, the ratio of feed to gain of groups B (1.51 ± 0.08) was 3.2% (P < 0.05) lower than that of group A (1.56 ± 0.11). This means that the L actobacillus ⅡCF2-1 fermented rice bran could improve the utilization rate of feed and promote their digestion and absorption.
The qualitative and quantitative analysis of intestinal microflora
By using the Illumina hiseq technology to completing 16S rRNA gene sequencing, high resolution characterization of the fecal bacterial microbiota under the 2 feeding treatments was measured and detailed bacterial profiles were obtained at genus precision for each group of samples.
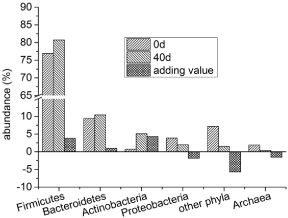
As Figure 4 shows, in the samples on the first day of feeding, the intestinal bacteria were dominated by Firmicutes (76.9%) and Bacteroidetes (9.4%), and their abundance accounted for 86.3%. In addition, it also contains Actinobacteria (0.7%), Proteobacteria (3.9%) and several other phyla (7.2% in total). Archaea accounts for 1.9%. After 40 days of feeding, the abundance of Firmicutes increased to 80.7 % (an increase of 3.8 %), Bacteroidetes increased to 10.4% (an increase of 1.0%), and Actinobacteria increased to 5.1% (an increase of 4.3%). Correspondingly, the abundance of the Proteobacteria and other phyla decreased to 2.0% (a decrease of 1.9%) and 1.5% (a decrease of 5.7%) respectively. The abundance of the Archaea also fell to 0.3% (a decrease of 1.6%).
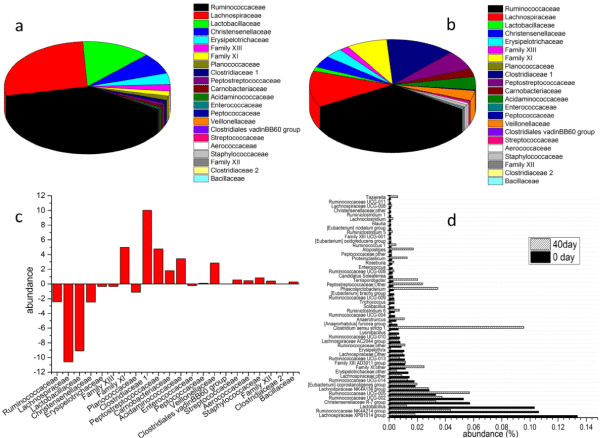
Ruminococcaceae, Lachnospiraceae, Lactobacillaceae and Christensenellaceae were the major families in Firmicutes at day 0, and their abundances were 28.8%, 21.1%, 10.3% and 5.9%, respectively (as Figure 5a shows). However, at day 40, the abundance of the four above were decreased obviously, while that of most of others families such as Family XI, Clostridiaceae 1, Peptostreptococcaceae and Acidaminococcaceae were significantly increased (Figure 5b and Figure 5c). At the genus level, the bacterial community were also obviously different between the samples of day 0 and day 40 (Figure 5d). In the fecal samples of 0 d, the major bacterial genera were Lachnospiraceae XPB1014 group (13.38%), Ruminococcaceae NK4A214 group (10.62%), Lactobacillus (10.33%), Christensenellaceae R-7 group (5.74%) and Ruminococcaceae UCG-002 (5.29%). But their abundances were decreased to 1.83%, 4.36%, 1.23%, 3.32% and 3.76% respectively at day 40. Many genera such as Ruminococcaceae UCG-005 (3.30 % to 5.71 %), Clostridium sensu stricto 1 (0.66 % to 9.56 %), Ruminiclostridium 6 (0.38% to 0.73%), Phascolarctobacterium (0.31% to 3.45%), Terrisporobacter (0.28% to 2.02%), their abundances were increased obviously. Although the abundance of Lactobacillus was decreased significantly, the number of the L. plantarumin was increased, as Table 3 shows. The quantity of L. plantarum increased significantly to 1.05 × 105 cfu/g (corresponding to 5.02 in table 3, p < 0.05) in the fecal samples of piglets fed the fermented rice bran, compared to that of fed basic diet (5.50 × 104 cfu/g, corresponding to 4.74 in table 3).
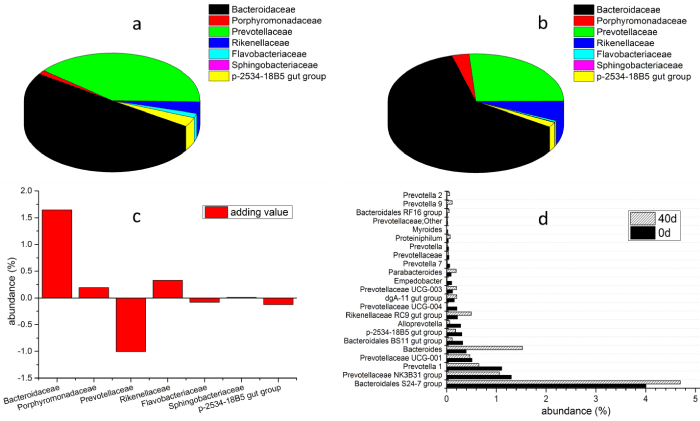
Bacteroidaceae and Prevotellaceae were the major families in Bacteroidetes both at day 0 and day 40 (Figure 6a and Figure 6b). However, the abundance of Bacteroidaceae was increased from 4.74% to 6.38% (increasing 1.64%, Figure 6c), while that of Prevotellaceae was decreased from 3.73% to 2.72% (decreasing 1.01%). In addition, the abundance of Rikenellaceae was increased from 0.37% to 0.71% (increasing 0.34%), and p-2534-18B5 gut group was decreased from 0.31% to 0.18% (decreasing 0.13%). Bacteroidales S24-7 (4.01% and 4.69%), Prevotellaceae NK3B31 (1.30% and 1.06%), Prevotella 1 (1.11% and 0.65%), Prevotellaceae UCG-001 (0.51% and 0.46%) and Bacteroides (0.40% and 1.52%) were the major genera in the bacterial community both in day 0 and day 40 (Figure 6d).
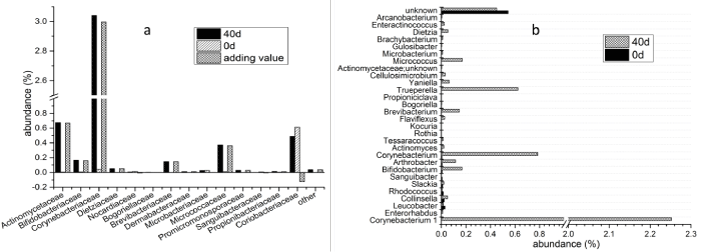
At day 0, Coriobacteriaceae was the major family in Actinobacteria, and its abundance was 0.61%, which account for about 83% of this phylum (Figure 7a), but it was decreased into 0.49% in the sample of day 40. However, the abundances of many other families such as Actinomycetaceae (from 0.01% to 0.68%), Bifidobacteriaceae (from 0.01% to 0.17%), Corynebacteriaceae (from 0.04% to 3.04%), Brevibacteriaceae (from 0.00% to 0.15%) and Micrococcaceae (from 0.01% to 0.37%), were increased significantly. An unknown genus in Coriobacteriaceae (abundance of 0.54%) was the major genus in Actinobacteria at day 0, and it was decreased to 0.41% at day 40 (Figure 7b). At day 40, Corynebacterium 1(2.25%), Corynebacterium (0.78%), Trueperella (0.62%) were the major genera. The abundances of others such as Bifidobacterium (0.01% to 0.17%), Micrococcus (0.00% to 0.17%), Brevibacterium (0.01% to 0.15%) and Arthrobacter (0.01% to 0.11%) were also increased compared with they were at day 0.
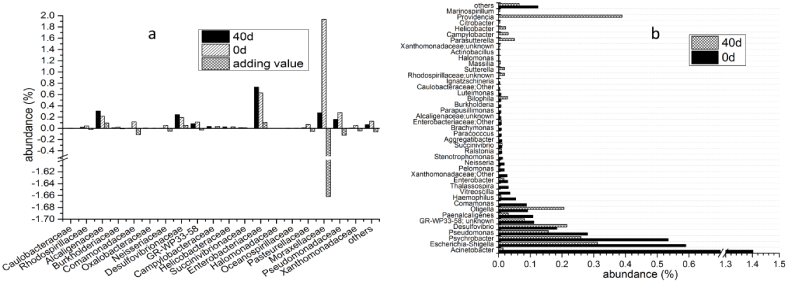
Moraxellaceae (1.94%) and Enterobacteriaceae (0.63%) were the major families in Proteobacteria at day 0 (Figure 8a). Most abundances of the families were decreased at day 40 and of Moraxellaceae was decreased to 0.27%. However, the abundance of Enterobacteriaceae were increased (to 0.73%). At genus level, Acinetobacter (1.40%), Escherichia-Shigella (0.59%) and Psychrobacter (0.53%) were the major genera at day 0 (Figure 8b). At day 40, abundances of most genera including many Pathogenic bacteria such as Enterobacter (0.03% to 0.02%) and Escherichia-Shigella (0.59% to 0.31%) were decreased significantly, which could potentially reduce the risk of colorectal cancer [21].
The Spirochaetae in the fecal samples including two genera, Sphaerochaeta and Treponema 2. Their abundance were also decreased from 0.05% to 0.02% and from 3.94% to 0.75%, respectively.
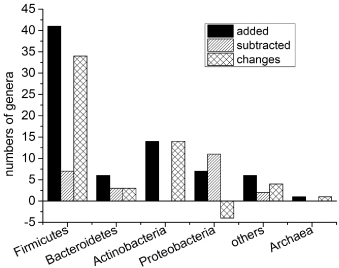
L. plantarum ⅡCF2-1 fermented rice bran also improved the microbiota biodiversity and altered microbiota community of growing pigs (Figure 9). A total of 75 genera were added which distributed at the statistical 6 phyla (including Archaea and others) and 23 were subtracted which distributed at 4 phyla at day 40 compared to day 0. Among them, the number of genera in Firmicutes was increased by 34 (41 added and7 subtracted), which is the most., followed by Actinobacteria of 14 (14 added). The largest decline in the number of genera was Proteobacteria, which was by -4 (7 added and 11 subtracted). It was the only phylum whose genera number has decreased.
Discussion
L. plantarum, a group of lactic acid bacteria that occur naturally in the natural fermentation cereal and silage, has been widely developed for commercial use as probiotics for animal [22]. Rice bran is rich in dietary fiber, which is potential sources of prebiotics for animal feed [23]. L .plantarum ⅡCF2-1 with advantages of rapid growth, short lag phase and high L-lactic acid yield has great potential in commercial feed application.
SSF is a simple technique for the production of bioactive compounds, which is economically viable due to the usage of agro-industrial residues, and also helps reduce the environmental impact of their disposal [24,25]. Compared to non-fermented rice bran, SSF of rice bran with L. plantarum ⅡCF2-1 had the higher nutritional and feeding value. L. plantarum could produce a variety of beneficial products, such as short chain fatty acid, protease, cellulase and so on [26], which may lead to the reduction of crude fiber and the alteration of protein's structure.
Trillions of microorganisms reside in the gastrointestinal tract of animals, most of which maintain a symbiotic relationship with their host, playing a critical part in biological processes such as nutrient utilization, resistance against infections, maturation of the immune system and host metabolism [26]. It was reported that the biodiversity of intestinal microbiota composition has the relationship to the improvement of feed efficiency [27,28]. In this study, the effects of added fermented rice bran was reflected directly in the significant improvement of performance of weaned piglets, including average daily gain, average daily feed intake and ratio of feed to gain. At the same time, the intestinal microorganisms of pigs also changed greatly before and after feeding fermented rice bran. There were 256 genera in the fecal sample at day 0, and the number was increased to 308 at day 40 (75 genera added and 23 subtracted). In addition, abundance differences at the genus level were also observed at day 40 compared to day 0. Among the 331 genera observed (including fecal samples at day 0 and at day 40), the abundance of 202 genera increased and of 29 genera decreased. The abundances of many beneficial bacterium such as Clostridium sensu stricto 1 (from 0.66% to 9.56%), Helcococcus (from 0.01% to 2.92%), Ruminococcaceae UCG-005 (from 3.30% to 5.71%) and Bifidobacterium (from 0.01% to 0.17%) were increased significantly. Abundance of many pathogenetic genera such as Enterobacter (from 0.03% to 0.02%) and Escherichia-Shigella (from 0.56% to 0.31%) were decreased.
The number of L. plantarum (1.05 × 105 cfu/g) at day 40 in fecal was increased for 2 times compared to day 0 (3.47 × 104 cfu/g). However, the abundance of Lactobacillus sp. decreased significantly. It indicated that the continuous probiotics intake could affect significantly piglets' microbiota, and the fermented rice bran improved the intestinal bacteria structure of piglets.
Conclusion
Rice bran fermented with L. plantarum ⅡCF2-1 could regulate the diversity and relative abundance of intestinal flora in growing pigs, as well as improve their growth and health performance.
Supplementary Materials
The following are available as file name of "Supplementary Materials", Table S1: Composition and nutrient levels of basal diets (air-dry basis).
Conflict of Interest
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Acknowledgments
This work was financially supported by Open Research Fund of Beijing Key Laboratory of Bioactive Substances and Functional Foods, Beijing Union University (No. SWHX202105), Grant Fund for Basic Science, Research of Central Nonprofit Academe (No. ZX2019) and Natl. Key R&D Program of China (No. 2018YFC1602703). The authors sincerely thank the SPF animal facility of Academy of National Food and Strategic Reserves Administration for the support of piglets feeding and intestinal of microflora analysis.
References
- Agaliya P, Jeevaratnam K (2012) Screening of Lactobacillus plantarum isolated from fermented Idli batter for probiotic properties. Afr J Biotechnol 11: 12856-12864.
- Chang M, Hong S, Chen J, et al. (2016) Antibacterial activity Lactobacillus plantarum isolated from fermented vegetables and investigation of the plantaricin genes. Afr J Microbiol Res 10: 796-803.
- Khemariya P, Singh S, Jaiswal N, et al. (2016) Isolation and identification of Lactobacillus plantarum fromvegetable samples. Food Biotechnol 30: 49-62.
- Valan Arasu M, Jung MW, Kim DH, et al. (2015) Identification and phylogenetic characterization of novel Lactobacillus plantarum species and their metabolite profiles in grass silage. Anna Microbio 65: 15-25.
- Wang J, Ji H, Zhang D, et al. (2011) Assessment of probiotic properties of Lactobacillus plantarum ZLP001 isolated from gastrointestinal tract of weaning pigs. J Biotechnol 10: 11303-11308.
- O'Shea CJ, Sweeney T, Bahar B, et al. (2012) Indices of gastrointestinal fermentation and manure emissions of growing-finishing pigs as influenced through singular or combined consumption of Lactobacillus plantarum and inulin. J Anim Sci 90: 3848-3857.
- Suo C, Yin Y, Wang X, et al. (2012) Effects of lactobacillus plantarumZJ316 on pig growth and pork quality. BMC Veterinary Res 8: 89.
- Pourali O, Asghari FS, Yoshida H (2010) Production of phenolic compounds from rice bran biomass under subcritical water conditions. Chem Engin J 160: 259-266.
- Abd Razak DL, Abd Rashid NY, Jamaluddin A, et al. (2017) Cosmeceutical potentials and bioactive compounds of rice bran fermented with single and mix culture of Aspergillus oryzae and Rhizopus oryzae. J Saudi Soc Agri Sci 16: 127-134.
- Kumar A, Kanwar S (2012) Lipase Production in Solid-State Fermentation (SSF): Recent developments and biotechnological applications. Dynam Biochem, Process Biotechnol Molecular Bio 6: 13-27.
- Katina K, Laitila A, Juvonen R, et al. (2007) Bran fermentation as a means to enhance technological properties and bioactivity of rye. Food Microbio 24: 175-186.
- Wang M, Lei M, Samina N, et al. (2020) Impact of Lactobacillus plantarum 423 fermentation on the antioxidant activity and flavor properties of rice bran and wheat bran. Food Chem 330: 127156.
- Kraler M, Schedle K, Domig KJ, et al. (2014) Effects of fermented and extruded wheat bran on total tract apparent digestibility of nutrients, minerals and energy in growing pigs. Anim Feed Sci Technol 197: 121-129.
- Zubaidah E, Nurcholis M, Wulan SN, et al. (2012) Comparative study on synbiotic effect of fermented rice bran by probiotic lactic acid bacteria lactobacillus casei and newly isolated Lactobacillus plantarum B2 in wistar rats. APCBEE Procedia 2: 170-177.
- Shibayama J, Goto M, Kuda T, et al. (2019) Effect of rice bran fermented with Saccharomyces cerevisiae and Lactobacillus plantarum on gut microbiome of mice fed high-sucrose diet. Beneficial Microbes 10: 811-821.
- Han W, Zhang XL, Wang DW, et al. (2013) Effects of microencapsulated Enterococcus fecalis CG1.0007 on growth performance, antioxidation activity, and intestinal microbiota in broiler chickens. J Anim Sci 91: 4374-4382.
- Amato KR, Yeoman CJ, Kent A, et al. (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. The ISME J 7: 1344-1353.
- Walter J, Tannock GW, Tilsala-Timisjarvi A, et al. (2000) Detection and identification of gastrointestinal lactobacillus species by using denaturing gradient gel electrophoresis and species-specific pcr primers. Appl Environ Microbiol 66: 297-303.
- Haarman M, Knol J (2006) Quantitative real-time PCR analysis of fecal lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol 72: 2359-2365.
- Zwietering MH, Jongenburger I, Rombouts FM, et al. (1990) Modeling of the bacterial growth curve. Appl Environ Microbiol 56: 1875-81.
- Yuan L, Zhang S, Li H, et al. (2018) The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed Pharmacother 108: 184-193.
- Kazun B, Malaczewska J, Kazun K, et al. Dietary supplementation with Lactobacillus plantarum and ß-glucan affects immune parameters in the tench (Tinca tinca) fry. Pol J Vet Sci.
- Zhuang X, Zhao C, Liu K, et al. (2018) Chapter 10 - Cereal grain fractions as potential sources of prebiotics: current status, opportunities, and potential applications. In: Ricke SC, Atungulu GG, Rainwater CE, et al. (editors). Food and feed safety systems and analysis: Academic Press. p. 173-91.
- Oliveira MdS, Feddern V, Kupski L, et al. (2010) Physico-chemical characterization of fermented rice bran biomass Caracterización fisico-química de la biomasa del salvado de arroz fermentado. CyTA - J Food 8: 229-236.
- Schmidt CG, Furlong EB (2012) Effect of particle size and ammonium sulfate concentration on rice bran fermentation with the fungus Rhizopus oryzae. Bioresource Tech 123: 36-41.
- El-Saadony MT, Alagawany M, Patra AK, et al. (2021) The functionality of probiotics in aquaculture: An overview. Fish Shellfish Immunol 117: 36-52.
- Quan J, Wu Z, Ye Y, et al. (2020) Metagenomic characterization of intestinal regions in pigs with contrasting feed efficiency. Front Microbio 11: 32.
- McCormack UM, Curião T, Buzoianu SG, et al. (2017) Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl Environ Microbiol 83: e00380- e00417.
Corresponding Author
Wei Han, Academy of National Food and Strategic Reserves Administration, Beijing 100037, PR China Tell: +86-010-58523596, Fax: +86-010-58523596 and Xuhui Zhuang, Academy of National Food and Strategic Reserves Administration, Beijing 100037, PR China Tell: +86-010-58523724, Fax: +86-010-58523724.
Copyright
© 2022 Liu Q. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.