Exogenous Salt and ABA Enhanced Stress Responsive Nuclear Protein in Solanum Lycopersicum
Abstract
Abiotic stresses such as salt and water stresses severely reduce crop productivity. Tomato (Solanum lycopersicum) is an important economic crop; however, not much is known about its stress responses. In our previous transcriptomic and computational studies we showed the level of salinity tolerance in different tomato cultivars. Rab genes were found quite responsive under salinity stress in two tomato cultivars (Pusa Ruby and Punjab Keshari). Here, putative promoter analysis of Rab11b was found to contain several stress related cis-elements.
To gain insight into stress-responsive gene regulation in tomato plants, we scanned the gene sequence of Rab11b gene from tomato database. The upstream region of which was then extracted. Rab protein family is one of the most important families in plant, plays an important role in the process of plant growth and development. Among several defence related cis-acting elements identified in the upstream region, MYB as an important ABA-responsive cis-element was selected and experiment of south-western analysis was aimed for studying the level of DNA-protein interaction.
Assays with nuclear extract show that the stress inducive protein factors can trans-activate tomato Rab11b gene by forming strong DNA-transcription factor complex in ABA and high salt treated tomato plants.
Keywords
ABA, MYB, MYC, ABRE, Lea/Rab gene, Abiotic stress, Salt stress, Tomato, Pusa Ruby, Punjab Keshari, Transcription factor, cis-elements
Introduction
Plant growth, development and productivity are severely affected by various environmental stresses including drought, salinity, extreme temperature, and heavy metals. Being sessile in nature, plants have to constantly encounter such stresses by altering the biochemical, physiological, and transcriptional activation of several stress-responsive genes [1-3].
Although remarkable advances have been made in recent times, the available information about mechanisms of plant responses to abiotic stresses in tomato plants is still scarce in comparison to Arabidopsis. Tomato is a member of the Solanaceae family and the third most commercially important crop. Cultivated tomato is overall considered as moderately tolerant to salinity [4]. Tomato was used as a model plant in several fields including genome sequence studies [5]. Global expression profiling is useful to understand the transcriptional regulation of stress-responsive genes for improving stress tolerance in tomato.
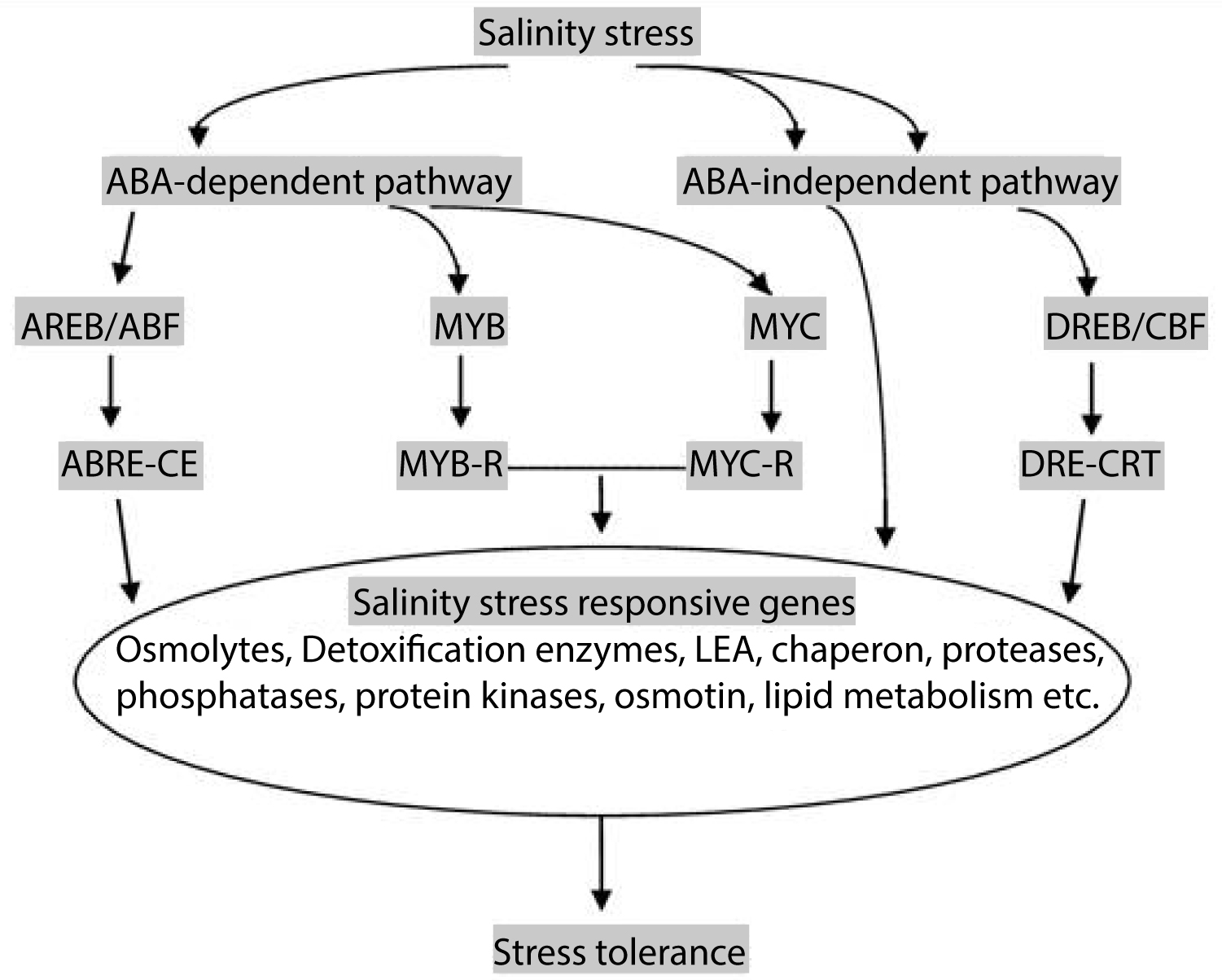
Responses to different abiotic stresses require the production of important metabolic proteins such as those involved in synthesis of osmoprotectants and regulatory proteins (Figure 1). In addition to such metabolic changes, a large set of plant genes commonly called lea (Late Embryogenesis Abundant) are transcriptionally activated, which leads to the accumulation of novel proteins in the vegetative tissues of plants under osmotic stress [6]. Lea/Rab proteins are essential for resurrection plants and seeds to protect the subcellular milieu against irreversible damage associated with adverse effect [7]. They are responsible for intracellular protein transport and also essential regulator of vesicle trafficking way [8,9]. Rab family is not only closely related to plant growth and development, but also plays an important role in stress and disease resistance.
Plant salt tolerance is a complex trait controlled by multiple genes. Transcription factors (TFs) are key regulatory proteins operating in signal transduction pathways. These regulatory proteins regulate gene expression by binding to specific DNA sequences in the promoters of respective target genes. This type of transcriptional regulatory system is called regulon. Several TFs targeting cis-acting elements involved in stress responsive gene regulation have been identified [10].
The expression of many lea proteins is regulated by abscisic acid (ABA). ABA is referred to as the plant stress hormone and many of the stress signals that activate transcription are controlled by this phytohormone ABA. ABA participates in the adaptation of plant species and tissues to osmotic stresses like drought and salinity [11]. Besides controlling stomatal aperture, ABA induces the expression of several genes with roles in dehydration tolerance in seeds and vegetative tissues [12-14]. There are existence of both ABA-dependent and ABA-independent pathways in the triggering of gene activation towards rapid stress response. Though the pathways can function independently but there are cross-talk mechanisms between them (Figure 1).
The promoters of such abiotic stress-responsive genes share regulatory sequences that are recognized by trans-acting factors participating in ABA signal transduction pathways. Among these sequences, the ABA-responsive element (ABRE) has been identified as the major cis-acting regulatory element in ABA-dependent gene expression [10], which is recognized by a specific group of basic leucine zipper (bZIP) transcription factors known as ABA-response element binding factors (AREBs) [15] or ABFs (ABRE-binding factors) [16]. TFs regulate a series of downstream stress-related genes, leading to biochemical and physiological modifications necessary for plant adaptation towards environmental perturbations [17,18].
Plants represent a large number of TFs which are classified by their DNA-binding domains [19]. The both ABA-dependent and ABA-independent signal transduction pathways from stress signal to expression of genes, involve different TFs such as AREB/ABF, MYC/MYB, DREB, NAM and their corresponding cis-regulatory elements ABRE, MYCRS/MYBRS, DRE, NACRS. In plants, MYBs having a wide range of function including the ABA-response, are well distributed and can work in conjunction with other transcription factors. A number of plant MYB TF members have been identified and characterized in numerous plant species Arabidopsis, apple, soyabean, maize , etc. [20-23]. The MYB TF family is not plant specific and is present in all eukaryotes. Extensive studies on MYB genes are there [24-27]. Based on the number of MYB domains, the MYB protein family has been classified into 4 different groups, such as 1R-, R2R3-, 3R- and 4RMYB proteins, respectively [25,26,28].
In our previous studies we showed the level of salinity tolerance in different tomato cultivars like Pusa Ruby, Punjab Keshari, Alisa Craig and Roma. Comparative transcript profiling [29] between Pusa Ruby (PR) and Punjab Keshari (PK) resulted in establishing PK as more salt tolerant variety than PR. A group of Lea genes (RabA, RabB, RabC, Rab11 were found generously responsive under salinity stress. In our another computational approach [30] towards investigating the molecular basis of high tolerance of tomato, we performed a comprehensive analysis of the proximal promoters of candidate members Rab1A, Rab1B, Rab1C, Rab11 of Rab gene family which are essential components of stress response. Analysis found the presence of various common stress related cis-acting elements (CREs) in the putative promoters of members of this gene family. These results brought about the idea on focussing into the stress related transcription factor binding with the upstream region of stress responsive plant genes.
Here we report the expression of transcription factor (putatively MYB related, not sequenced) which has been found to be activated by NaCl/ABA induction on two different tomato cultivars (Pusa Ruby and Punjab Keshari). Subcellular localization assay indicated this factor as a nuclear-localized protein with transcriptional activity by binding to the Rab11b promoter sequence bearing the MYB consensus DNA motif (CNGTTR).
Materials and Methods
Plant materials, Plant growth and stress treatments
Seeds of Solanum lycopersicum (L.) Pusa Ruby cv and Punjab Keshari cv, were kindly provided by the Amtala Seed Center, Amtala, West Bengal. Plants were grown in controlled environment chambers at 25-26 ℃, 50% relative humidity, with 8-hour photoperiod. For comparison, two types of tomato cultivars, Pusa Ruby (PR) and Punjab Keshari (PK) were cultured in a hydroponic system (Figure 2) with periodical replacement with fresh media. Seedlings were grown for 2 months in half strength MS [31] solution. For salt treatment, eight-week-old plants in a tray containing 0.5 × strength MS solution were supplemented with 0/200 mM NaCl (6 h) and 25 μM ABA (6 h and O/N). Then, the plants were washed thoroughly with sterile de-ionized water, their leaves were sampled separately and stored at -80 ℃ for investigations. Three replicates were performed for each treatment.
Preparation of tomato nuclear extract and South-Western Blot with the DNA probe harboring MYB motifs of Rab11b upstream
Nuclei were prepared from leaf samples of 2-month-old tomato plants according to the method of Roychoudhury, et al. [32] with some modifications. All steps were performed at 0 to 4 ℃. Plant tissues were ground to a fine powder with liquid N2 Tomato nuclear extract was prepared as described by Oeda, et al. [33] with minor changes, and the protein content estimated by Bradford method [34].
About 20 pmoles (220-240 ng) of each of the two synthetic oligonucleotides were labeled at the 5' end with 100 μCi of [γ32P]-ATP (5,000 Ci mmol-1), the two complementary phosphorylated oligonucleotides (Table 1) were mixed together and annealed by slow cooling, followed by probe purification. Binding reactions were performed essentially as described by Oeda, et al. [33].
South Western Blot Analysis was performed according to Roychoudhury, et al. [32] with slight modifications. About 50 μg of nuclear proteins were boiled with standard SDS buffer and separated by 10% SDS-polyacrylamide gel and then electrophoretically transferred onto PVDF membrane (Hybond-P from Amersham Pharmacia). After blocking the membrane with 100 μg ml-1 sonicated salmon sperm DNA (Amersham Pharmacia), [γ32P]-ATP DNA probe (105 CPM ml-1 count) was added to the blot and incubated overnight at room temperature (25 ℃) with constant shaking in 1x binding buffer (10 mM Tris-Cl pH 7.5, 50 mM NaCl, 1 mM EDTA, 5 mM MgCl2). The blot was washed 4-5 times with increasing concentration of salt from 50 to 150 mM, semidried and then exposed to Kodak X-Omat film for autoradiograph.
Computational analysis
The SGN (Sol Genomic Network) PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; [35] RefSeq NCBI (http://www.ncbi.nlm.nih.gov). were used for gene sequences extraction. PLACE and PlantPAN were used to scan the cis-elements, in the putative Rab11b promoter sequence. The significance of candidate cis-elements (MYB, MYC, DPB, WRKY, GATA) was evaluated through scanning further with the plant promoter analysis databases, PLACE (http://www.dna.affrc.go.jp/PLACE/; [36] and PlantPAN web tool (PlantPAN; http://PlantPAN.itps.ncku.edu.tw3.0).
Results
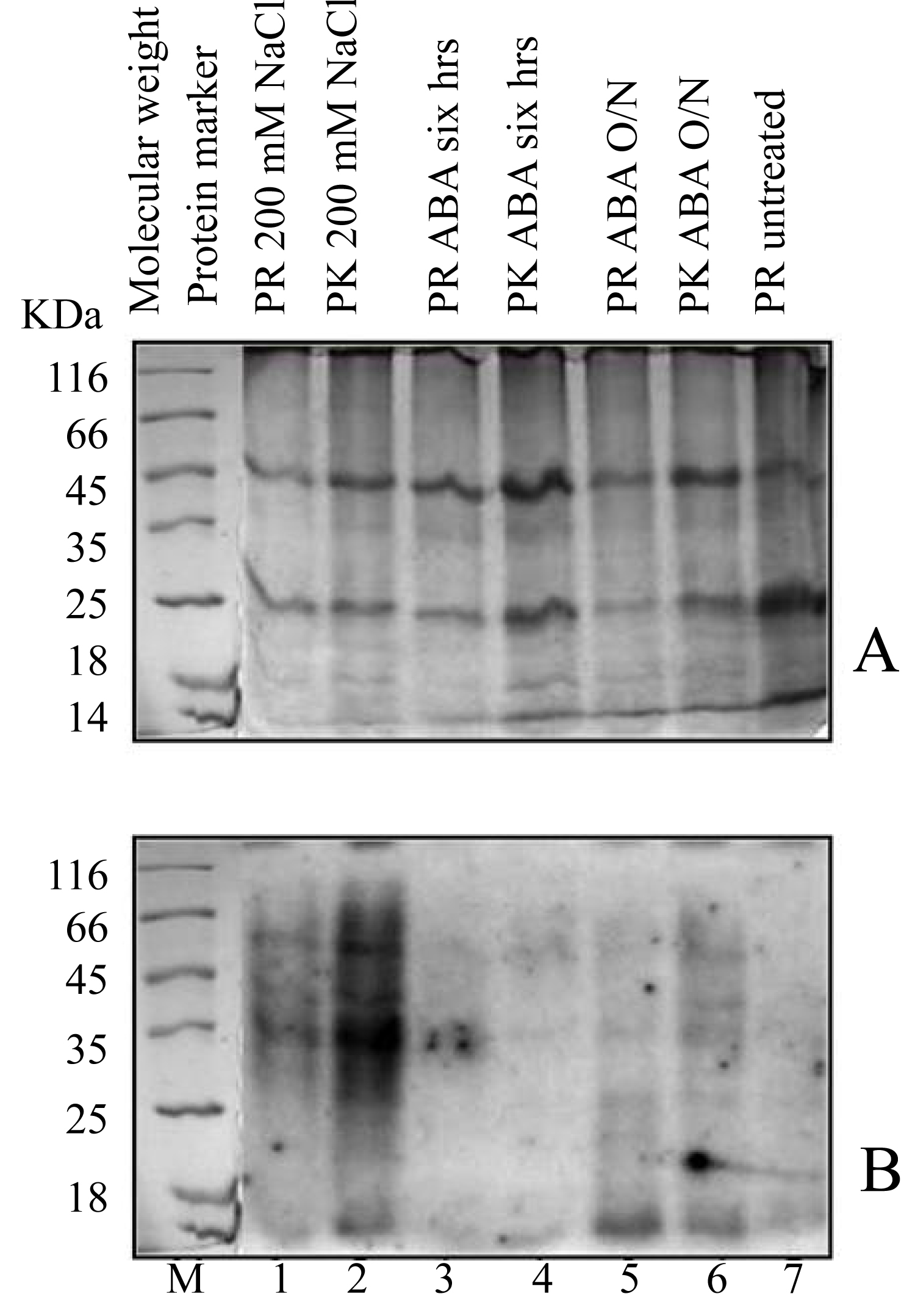
Tomato database search revealed that the putative promoter region (Supplementary 1) host a number of cis-elements and some of them are known to be abiotic stress responsive. Here our experiment with double stranded oligonucleotide probe containing MYB motifs, was found to be interacted with unknown protein factors under salinity stress and ABA imposition. In NaCl treated leaves this DNA-protein complex formation was much stronger than ABA treated leaves. A comparison between the two tomato cultivars shows differential response to stress (Figure 2 and Figure 3).
Transcription factors binding to the MYB motif containing DNA probe that responds to salinity stress and ABA treatment in tomato plants
Southwestern blot analysis identified a ∼55 and ∼35 kDa transcription factors (Figure 3) in the nuclear extract of all treated samples of tomato (NaCl or ABA). The intensity of the 35 kDa protein was sharper under salinity stress than in ABA treated leaf samples of both the cultivars, thereby clearly suggesting a role for this protein in salinity stress response mediated by ABA (Figure 3). Responses were found somewhat similar in ABA treated samples of both the cultivars but MYB-like cis element binding complex formation was much distinct in NaCl treated PK samples (Figure 3). Thus the presence of a tomato factor that can trans-activate tomato Rab11b gene was confirmed triggering to the fact that Rab11b gene is very likely to show expression under its own promoter when subjected to stressful condition in tomato. So this South-western blot result is consistent with the results obtained through previous transcriptomic and other analysis [29,30].
Discussion
Transcriptional Regulation of lea genes is mainly operated through ABA. This stress responsive hormone regulates several aspects of plant development, including seed development, seed dormancy and desiccation tolerance of seeds. Abiotic stress signal network and gene expression involve different cis and trans-acting elements. The basic leucine zipper factors, AREB/ABF and MYC/MYB proteins are mainly responsible for activating major ABA-dependent stress response through different ABREs and MYBRS and MYCRS. MYB TFs have been deeply studied to play a regulatory role in a stressful environment and constitutes one of the largest TF groups that are functionally diverse and found in all eukaryotes.
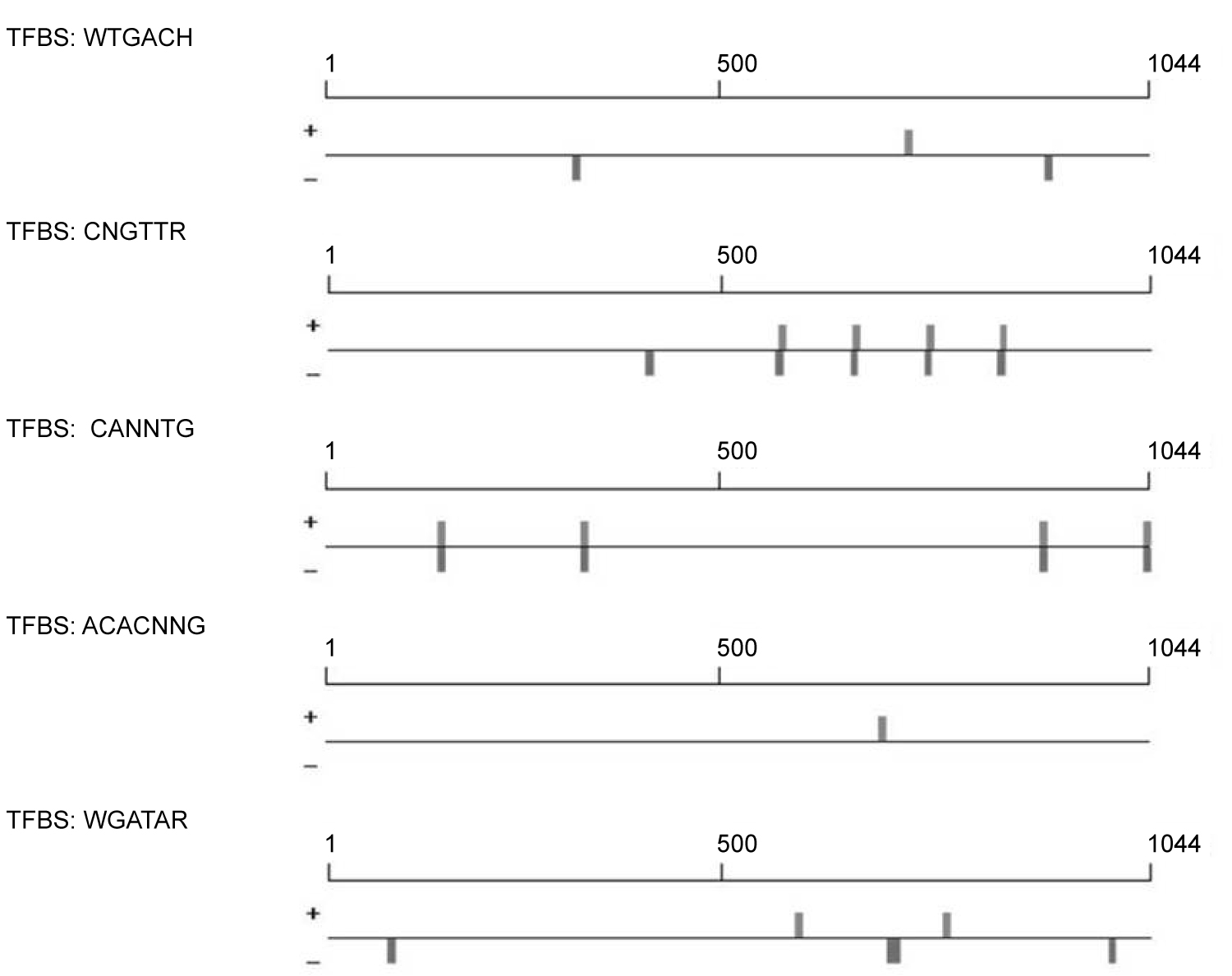
In one of our previous experiments, expression of Rab11a/b [29] was found to be increased with high salinity stresses in tomato plants. PK tomato cultivar was found as more tolerant than PR. The strong induction of Rab11a/b by salinity/abiotic stresses prompted us to read its promoter sequence (1,000 bp upstream the transcription start site) by searching the prompter sequence against the PLACE database (http://www.dna.affrc.go.jp/PLACE/). The promoter sequence indeed hosts many putative stress response-related cis-elements, such as WRKY (3 hits), DPB (1 hit), MYC recognition site (8 hits), MYB recognition site (9 hits), and GATA box (5 hits) (Figure 4). Results based on south-western blot, clearly suggesting a role of transcription factors (expectedly MYB-like) in salt stress response. Intensity of binding is sharpest in PK cultivar (showing its supremacy over PR) when treated with NaCl. Another intriguing observation is that this DNA-protein binding was induced by ABA even in the untreated plants. All these are suggesting a role of this protein in abiotic and salinity stress. The result also agreed with the findings previously reported [29,30].
A fair amount of previous studies demonstrate that regulation of gene expression at the transcriptional level is primarily controlled by a promoter and harboring a number of cis acting regulatory elements [37,38]. It is also evidenced that, yeast one-hybrid technology is a classical method for studying the interaction between DNA and protein, and allows the effective isolation and identification of proteins that bind to specific DNA sequence [39,40].
The expression of lea genes is mediated by the transcription factors which activate CRT, ABRE and MYC/MYB cis-elements through the signaling network of Ca2+ and ABA in plants under salinity stress. However, the ABA dependent and ABA-independent pathways act in parallel and also interact, thereby providing added coordination between stress signals and ABA in the regulation of stress-responsive genes [41,42].
However, the basic functional architecture for molecular responses, interactions and cross talk towards abiotic stress needs to be studied. Study of large number of ABA-dependent and independent transcription factors and their downstream binding elements can give better understanding of the molecular basis of stress tolerance, which involves complex network of genes operating in co-ordination.
The Arabidopsis homolog of SlAREB is AREB1, which is a well-known transcription regulator in the ABA-mediated stress signal transduction pathway [15]. bZIP proteins characteristically contain a highly conserved bZIP domain, which is composed of a basic region for sequence-specific DNA binding and a leucine zipper for dimerization specificity [43]. Genetic and molecular studies suggest that bZIP TFs function in seed maturation and germination, ABA and/or stress signaling, and defense and photomorphogenesis [43-49]. Though ABRE is the major cis-element for ABA-responsive gene expression [10], but strong binding of TF with MYB element present in Rab11b promoter has a role in stress tolerance. So far 127 MYB genes have been identified in tomato. Many such genes were reported to be involved in abiotic stresses. A recent research [50] demonstrated that in Brachypodium distachyon, several MYB genes were up-regulated in presence of ABA and also in response to stresses like salinity or draught. Abe, et al. [51] showed that the Arabidopsis MYB transcription factor proteins AtMYC2 and AtMYB2 function as transcriptional activators in ABA-inducible gene expression, suggesting a novel regulatory system for gene expression in response to ABA, other than the ABRE-bZIP regulatory system. Future identification and molecular characterization of this factor will be helpful for clarifying the mechanisms behind this DNA binding association conferring salt tolerance in tomato cultivars in the present study.
Conclusion
Tomato is one of the most widely cultivated vegetable crop species and more research needed to improve the current level of understanding of the physiological and molecular mechanisms related to stress response. Here, a comparison was made between two tomato cultivars contrasting with respect to their salinity tolerance. Study we targeted the MYB like consensus sequence (motif-5′-CNGTTR-3′) on the candidate Rab11b promoter potentially involved in response to salt stress. Southwestern blotting has been used to investigate DNA-protein interactions and subsequent prediction of potential protein factors. Analysis provides the information regarding its size and binding intensities. This analysis revealed the very involvement of putative MYB like factors in transcriptionally co-regulatory networks and actively functional under abiotic stress condition. It still remains ill-defined what the underlying complex mechanism behind such response of the factor to salinity stress might be. Addressing these questions is the aim of the continuation of this study further.
Acknowledgement
The author is thankful to Prof. Uday Banerjee, Director of the Bose Institute. Thanks also go to the Chairperson of the Division of Plant Biology (DPB). The author acknowledges the support of the Central Instrument Facility of the Bose Institute.
References
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways, Curr Opin Plant Biol 3: 217-223.
- Li T, Sun JK, Bi YP, et al. (2016) Overexpression of an MYB-related gene FvMYB1 from fraxinus velutina increases tolerance to salt stress in transgenic tobacco. Journal of Plant Growth Regulation 35: 632-645.
- Hu JT, Chen GP, Yin WC, et al. (2017) Silencing of SlHB2 improves drought, salt stress tolerance, and induces stress-related gene expression in tomato. Journal of Plant Growth Regulation 36: 578-589.
- Mueller LA, Tanksley SD, Giovannoni JJ, et al. (2005) The tomato sequencing project, the first cornerstone of the international Solanaceae project (SOL). Comp Funct Genomics 6: 153-158.
- Pérez-Alfocea F, Balibrea ME, Santa Cruz A, et al. (1996) Agronomical and physiological characterization of salinity tolerance in a commercial tomato hybrid. Plant and Soil 180: 251-257.
- Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2: 503-512.
- Silva Artur MA, Rienstra J, Dennis TJ, et al. (2019) Structural plasticity of intrinsically disordered LEA proteins from Xerophyta schlechteri provides protection in vitro and in vivo. Front Plant Sci 10: 1272.
- Novick P, Zerial M (1997) The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol 9: 496-504.
- Brennwald P (2000) Reversal of fortune: Do Rab GTPases act on the target membrane? J Cell Biol 149: 1-3.
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221-227.
- Verslues PE, Zhu JK (2005) Before and beyond ABA: Upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33: 375-379.
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52: 1569-1582.
- Finkelstein RR (2013) Abscisic acid synthesis and response. Arabidopsis Book 11: e0166.
- Li K, Xing C, Yao Z, et al. (2017) PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol J 15: 1186-1203.
- Uno Y, Furihata T, Abe H, et al. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A 97: 11632-11637.
- Choi H, Hong J, Ha J, et al. (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723-1730.
- Ouyang B, Yang T, Li HX, et al. (2007) Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot 58: 507-520.
- Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: Molecular regulation and stress responses in plants. Front Plant Sci 7: 760.
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447-456.
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423-434.
- Cao ZH, Zhang SZ, Wang RK, et al. (2013) Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS One 8: e69955.
- Du H, Feng BR, Yang SS, et al. (2012) The R2R3-MYB transcription factor gene family in maize. PLoS One 7: e37463.
- Li Q, Zhang C, Li J, et al. (2012) Genome-wide identification and characterization of R2R3 MYB family in Cucumis sativus. PLoS One 7: e47576.
- Du H, Zhang L, Liu L, et al. (2009) Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Mosc) 74: 1-11.
- Dubos C, Stracke R, Grotewold E, et al. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573-581.
- Ambawat S, Sharma P, Yadav NR, et al. (2013) MYB transcription factor genes as regulators for plant responses: An overview. Physiol Mol Biol Plants 19: 307-321.
- Zhenjun Li, Rihe Peng, Yongsheng Tian, et al. (2016) Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol 57: 1657-1677.
- Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J Mol Evol 46: 74-83.
- Roy C (2015) A comparative molecular analysis of two genotypes of Solanum lycopersicum in respect of the expression of selected salt responsive genes in leaves and roots. BioTechnologia 96: 307-316.
- Roy C (2017) Computational approaches to identify regulators of stress inducible genes of rab family in S. lycopersicum. Genetics and Molecular Biology 7: 99-110.
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473-497.
- Roychoudhury A, Roy C, Sengupta DN (2007) Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep 10: 299-371.
- Oeda K, Salinas J, Chua NH (1991) A tobacco bZIP transcription activator (TAF-1) binds to a G-box-like motif conserved in plant genes. EMBO J 10: 1793-1802.
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Lescot M, Déhais P, Thijs G, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325-327.
- K Higo, Y Ugawa, M Iwamoto, et al. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297-300.
- Mithra SVA, Kulkarni K, Srinivasan R (2017) Plant promoters: Characterization and applications in transgenic technology. Plant Biotechnology: Principles and Applications, 117-175.
- Zhang L, Song Z, Li F, et al. (2019) The specific MYB binding sites bound by TaMYB in the GAPCp2/3 promoters are involved in the drought stress response in wheat. BMC Plant Biol 6: 366.
- Shu-Ying F, Kazuhisa O, Takashi I (2010) A yeast one-hybrid system to screen for methylated DNA-binding proteins. Nucleic Acids Res 38: e189.
- Bonaldi K, Li Z, Kang SE, et al. (2017) Novel cell surface luciferase reporter for high-throughput yeast one-hybrid screens. Nucleic Acids Res 45: e157.
- Lan Thi Hoang X, Du Nhi NH, Binh Anh Thu N, et al. (2017) Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr Genomics 18: 483-497.
- Maheshwari P, Divya Kummari, Sudhakar Reddy Palakolanu, et al. (2019) Genome-wide identification and expression profile analysis of nuclear factor Y family genes in Sorghum bicolor L. (Moench). PLoS One 14: e0222203.
- Nijhawan A, Jain M, Tyagi AK, et al. (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333-350.
- Jakoby M, Weisshaar B, Droge-Laser W, et al. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106-111.
- Lopez-Molina L, Mongrand S, McLachlin DT, et al. (2002) ABI5 acts downstream of ABI3 to execute an ABA dependent growth arrest during germination. Plant J 32: 317-328.
- Zhang Y, Tessaro MJ, Lassner M, et al. (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647-2653.
- Fujita Y, Fujita M, Satoh R, et al. (2005) AREB1 Is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought Stress tolerance in Arabidopsis. Plant Cell 17: 3470-3488.
- Thurow C, Schiermeyer A, Krawczyk S, et al. (2005) Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J 44: 100-113.
- Mallappa C, Yadav V, Negi P, et al. (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J Biol Chem 281: 22190-22199.
- Abe H, Urao T, Ito T, et al. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in Abscisic acid signaling. Plant Cell 15: 63-78.
- Chen S, Niu X, Guan Y, et al. (2017) Genome-wide analysis and expression profiles of the MYB genes in Brachypodium Distachyon. Plant and Cell Physiol 58: 1777-1788.
Corresponding Author
Chaitali Roy, Division of Plant Biology, Bose Institute, Block EN, Sector-V, Salt Lake, Kolkata-700091, India, E-mail: chaitalli.bi@jcbose.ac.in
Copyright
© 2020 Roy C. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.