Expression of Cucumber Green Mottle Mosaic Virus Movement Protein in Cucumber Leads to the Expression Changes of Endogenous Gene
Abstract
Cucumber green mottle mosaic virus (CGMMV) is one of the most important diseases of cucurbit crops. To date the only method available to control this devastating disease is the use of resistant varieties or disease-resistant rootstocks. However, the development of transgenic technology offers the potential to create resistant varieties through the expression of foreign genes. Such approaches are not without risk, and it has been noted that introduction of transgenes can have wide ranging effects, often affecting non-target processes. The current study was therefore initiated to investigate the effect of genetic modification on 12 related genes in transgenic cucumber seedlings expressing the CGMMV movement protein (CGMMV-MP) at the two-true-leaf stage. Compared with non-transgenic cucumbers (cv. Zhongnong 16), the results of quantitative PCR (qPCR) indicated that six of the genes had significant altered expression in the transgenic plants, four that were up-regulated including the cucumber peeling cupredoxin, Histone H4, Cytochrome oxidase and Thaumatin-like protein and two that were down-regulated, cytochrome b6-f complex and disulfide isomerase. The data collected therefore provide greater understanding of the impact of introduced exogenous genes in cucumber, as well as highlighting resistance genes that have the potential to prevent CGMMV infection.
Keywords
Cucumber green mottle mosaic virus, Yeast two-hybrid system, Transgenic cucumber, qPCR
Abbreviations
PCR: Polymerase China Reaction; qPCR: Quantitative Real-time Polymerase China Reaction; CGMMV: Cucumber Green Mottle Mosaic Virus; MP: Movement Proteins; Bt: Bacillus Thuringiensis; SEM: Scanning Electron Microscope; YTHS: Yeast Two-Hybrid System; iTRAQ: Isobaric Tags for Relative and Absolute Quantitation; BA: 6-Benzyladenine; MS: Murashige and Skoog; IAA: Indole-3-Acetic Acid; CB: Carbenicillin; NAA: Neomycin Phosphotransferase; Kan+: Kanamycin
Introduction
Cucumber green mottle mosaic virus (CGMMV), which belongs to the Tobamovirus genus of the Virgaviridae family, was first reported in Cucumis sativus from Great Britain [1], but has quickly spread to most regions of the world [1-9]. As well as being soil borne, the disease can be spread by contaminated plant materials, including seeds, pollen and vegetative propagation stock, and is easily transmitted to healthy cucumber plants [7,10,11]. Although precautions can be taken to avoid the spread of CGMMV between crops and different geographic regions, once CGMMV has been introduced to fields or nurseries, all infected plants, as well as suspect plants from the surrounding area, must be removed and destroyed [12]. In the absence of effective methods of control, CGMMV, which has a wide host range, has become one of the most devastating pathogens of cucurbitaceous crops. However, recent developments using transgenic plants have shown that expressing components of CGMMV genome, including the coat protein (CP), movement protein (MP) and RNA replicase, can induce CGMMV resistance in cucumber plants via post-transcriptional gene silencing [13,14]. Such CGMMV-resistant varieties could be an invaluable tool for control CGMMV during seed production or the preparation of vegetative propagation stocks by grafting.
The genomes of most plant viruses contain genes that encode movement proteins (MP), which facilitate the movement of virus particles between cells via the plasmodesmata (PD), but also systemically through the entire plants via the phloem sieve tubes [15]. However, despite having an essential role in the proliferation of viral diseases, it has also been noted that MPs can be key a virulence factors mediating resistance against virus infection [13]. Moreover, there is growing evidence that the transgenic expression of MP genes can restrict the spread of plant viruses via RNA silencing [16]. However, studies have also shown that the expression of such transgenes can cause unintended effects altering the symptoms of non-target viruses resulting in hybrid viral infections [17]. In addition, it is well established that transgenes can cause changes to the morphology, physiology and metabolism of the recipient plants [18]. For example, expression of the MP gene of potato leafroll virus (PLRV) in tobacco plants resulted in reduced rates of photosynthesis and higher carbohydrate content [19]. It has also been found that the transgenes in genetically modified (GM) plants can affect processes such as seed dispersal, cross-pollination and the production of secondary metabolites and thereby affect their surrounding environment. A well-known example of this is the transgenic cotton (Gossypium hirsutum L.) that expresses the endotoxin gene from Bacillus thuringiensis (Bt) originally developed to control insects pests [20]. However, subsequent research has revealed that many variables including, enzyme activities and mineral-N (NH4+-N+NO3--N) were significantly reduced in fields planted with Bt cotton compared with non-Bt cotton [21]. Nonetheless, it is widely accepted that transgenic crops have great potential to increase crop production, even though some controversy still remains about how these foods should be regulated [22]. The use of viral genes to induce resistance in transgenic plants has great potential for the management of viral diseases although some doubts still remain about their safety [23]. The current study was therefore initiated to evaluate the expression level of genes in transgenic cucumber plants expressing the CGMMV movement protein (CGMMV-MP) in an attempt to provide sufficient data on the safety of the plants before larger scale field trials. The target genes, which were identified, using the yeast two-hybrid system (YTHS) and the results from iTRAQ screening, were assessed their expression levels in the transgenic cucumber plants expressing the CGMMV-MP.
Materials and Methods
SEM and RT-PCR confirming CGMMV infection of inoculated leaves
The CGMMV inoculum was obtained from the Chinese Academy of Inspection and Quarantine (CAIQ), Beijing, China, and maintained according to the published protocol [24]. The leaves from inoculated cucumber plants (cv. Zhongnong 16) were confirmed to be infected with CGMMV using SEM and RT-PCR. The suspected contaminated cucumber leaves were ground in 0.1 M phosphate buffer with pestle and mortar [25]. Then, adopted the scanning electron microscope (SEM) to observe the CGMMV particles in 5 μL plant extracts, the procedures for SEM were followed as for the previous studies [24].
The total RNA was extracted from 100 mg leaf using the EASYspin Kit (Biomed, Beijing, China). The RT-PCR was performed on MyCycler thermo cycler (Bio-Rad). For the PCR reaction, 1 μL of RNA was added into 13 μL of the PCR reaction mixture containing 12.5 μL of 2 × reaction mix and 0.5 μL of SuperScript® III RT/Platium® taq high fidelity enzyme mix (Invitrogen, U.S.), 0.5 μL of 5 μM primers PMU, 5'-actgagctcctaggtgtgatcggattgta-3' and PMD, 5'-gactctagatagtctctaagtaaggtgtcag-3', that were designed against the sequence contained in the NCBI data base (Accession No. D12505). Finally, added DEPC treated water to make 25 μL reaction volumes. The primers contained SacI and XbaI restriction sites, respectively. The PCR protocols was as following: 50°C for 30 min, 95°C for 2 min, 94°C for 30s, 55°C for 30s, 68°C for 1 min, total 35 cycles was running and then annealing for 10 min at 68°C. The PCR products were visualized using electrophoresis on a 1% agarose gel. The PCR products were recovered using a gel extraction kit (Biomed), cloned into the pEASY-T3 vector (TransGen Biotech) and sequenced (Biomed, Beijing, China). The sequences were compared to CGMMV sequences in the National Center for Biotechnology Information (NCBI) database using a blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Screening infected cucumbers for proteins associated with the CGMMV-MP using the YTHS. Total RNA was extracted from cucumber's leaves (cv. Zhongnong 16). The 2 × 106 cfu/ml cDNA libraries in Escherichia coli DH5α were prepared using the SMART cDNA library construction kit (TaKaRa, Dalian, China) according to the protocol of the manufacturer. The pGBKT7 and pGADT7 vectors, as well as the two yeast strains AH109 and Y187, were kindly donated by Wang Tao (Life sciences center, China Agricultural University, Beijing, China).
The YTHS analysis was conducted in accordance with the protocol of a previous study [26]. The MP gene from CGMMV was first amplified using the gene specific primers: Fmp, 5'-gacgaattcatgtctctaagtaaggtgtcag-3' and Rmp, 5'-atcctgcagctaggtgtgatcggattgta-3', which contained EcoRI and PstI restriction sites, respectively, and cloned into pGKBT7, while the cDNA library was constructed in pGADT7. The vectors were then transformed into the competent yeast cells of AH109 and Y187, respectively. After establishing that the vectors had no toxicity, the two yeast strains were mixed and allowed to hybridize in YDPA medium (Kan+, 50 μg/mL), before being selected on auxotrophic media (SD/-Leu-Trp-His-Ade). Protein-protein interactions were detected using the LacZ method described in a previous study [27]. There are 10 positive colonies with blue color of each protein were picked and used for sequencing. Also, these positive colonies were screened with auxotrophic media (SD/-Leu-Trp-His-Ade) to confirm them whether grow on or not after the pGADT7-cDNA co-transform with pGKBT7-CGMMV-mp.
Agrobacterium-mediated transformation of cucumber seedlings
The movement protein was amplified by primers PMU and PMD. The resulting Ti Plasmid, a derivative of pBi121 (The key laboratory of plant pathology, China Agricultural University, China), was then inserted into Agrobacterium tumefaciens (C58C1), which was used to transfection cucumber explants as follows.
Cucumber seeds (cv. Zhongnong 16) were first soaked in distilled water for 30 minutes before being surface sterilized with sodium hypochlorite (NaOCl, 1%) for 5 minutes. The seeds were then washed with distilled water before being cultured for two days on MS medium at 26°C with a (16 h/d) dark-light cycle until they germinated and thereafter during light and dark culture until the seedlings produced buds. Sections were then cut from the meristem at the first-true-leaf stage and dark-incubated on MS medium (BA, 0.8 mg/L and IAA, 0.2 mg/L) at 26°C for 2 days. The explants were then soaked in an Agrobacterium (bearing the recombinant vector pBI121 containing the MP gene) suspension that contained the BA and IAA (0.8 mg/L and 0.2 mg/L, respectively) for 10 minutes before being transferred to MS medium supplemented with AS (1 μL/mL). The explants were placed on top of sterilized filter paper that had been placed on the sold medium and dark incubated at 26°C. After two days the explants were transferred to fresh MS medium containing BA, IAA, Kan+ (25 mg/L) and CB (500 mg/L) and light-incubated at 26°C. The explants were transferred to fresh medium every two weeks until buds developed. At this point the explants were transferred to MS medium containing NAA (0.1 mg/L), Kan+ and CB and maintained in a similar fashion until roots developed.
Estimating the expression level of CGMMV-MP associated proteins using qPCR
The expression levels of 12 related genes, six identified by YTHS (Table 1) and eight by iTRAQ analysis (Table 1), which were obtained from the previous research [28], were assessed by qPCR (the used primer are list in table 2). For the selected genes, either interactions with CGMMV-mp or causing by CGMMV infection, which including significantly change expressing level or its characteristic has been confirmed with pathogenesis-related in previous studies. Total RNA was extracted from both transgenic and wild-type (cv. Zhongnong 16) seedlings at the two-true-leaf stage using 3 μg RNA and a cDNA synthesis kit (Invitrogen, U.S.) according to the protocol of the manufacturer. The qPCR itself was conducted in 25 μl reaction mixtures containing 1.0 μl cDNA, 0.5 μl each of the forward and reverse primers (10 μM), 12.5 μl 2 × TaqMix, and 1.25 μl eva green, and processed by real-time PCR using the T100 Thermal cycler (BIO-RAD, U.S.) and the following program: denaturation at 95°C for 5 mins followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 20 s, and extension at 72°C for 20 s. qPCR detection was repeated three times along with three independent repetitions. A Student's t test was used to determine statistical significance.
Results
Confirmation of samples and the selection of pathogenesis-related proteins
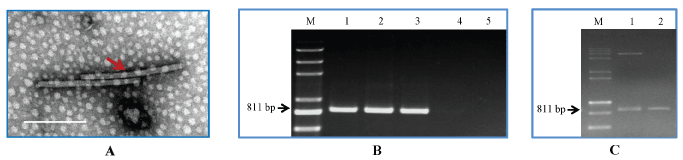
Leaf samples were confirmed to be infected with CGMMV by SEM and qPCR (Figure 1A and Figure 1B) before being used to clone the CGMMV-MP gene (Figure 1C) in the pGKBT7 vector and construct the cDNA libraries in pGADT7, respectively. The CGMMV particles displayed about 300 nm of length in figure 1. The hybridization solution containing the two strains was then plated on selective medium (SD/-Leu/-Trp/-Ade/-His) to screen proteins that interacted with the CGMMV-MP. After against the sequences with the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), a total of six genes were identified including several typical pathogenesis-related proteins (Table 1). It was interesting to note that the YTHS analysis identified two proteins, cytochrome b6-f complex and a thaumatin-like protein, which had been among the eight pathogenesis-related proteins (Table 1) identified by a previous iTRAQ study [28].
Expression levels of related genes in transgenic cucumber containing the CGMMV-MP
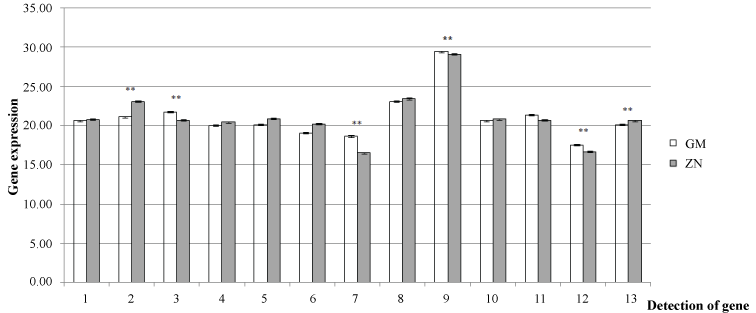
The MP gene (Appendix 1) of CGMMV was cloned into the T-DNA of the pBi121 Ti Plasmid and used to transform cucumber seedlings by Agrobacterium-mediated transformation. The expression levels of the six interaction proteins identified by the YTHS analysis and six pathogenesis-related proteins identified using iTRAQ analysis [28] were assessed by real-time PCR (Table 1 and Table 2). Tubulin (Accssion No.AJ715498 was from NCBI database) was chosen as a reference gene. Twenty-five independent transgenic seedlings were generated from different cucumber callus. Extracted the total RNA from the leaf of different seedlings and then evaluated 12 related genes expressing in 25 transgenic cucumbers, respectively. The results indicated that six of the genes evaluated had significantly altered expression compared to healthy cucumber (Figure 2), four that were up-regulated including cucumber peeling cupredoxin, histone H4, cytochrome oxidase and the thaumatin-like protein and two that were down-regulated including cytochrome b6-f complex and disulfide isomerase.
Discussion
Twelve related proteins were identified in the current YTHS analysis and previous iTRAQ study, were selected to investigate how the transgenic expression of the CGMMV-MP in cucumber seedlings affected the expression levels of endogenous genes. Only six of the genes assessed were found to have significant altered expression in the genetically modified cucumber seedlings. The thaumatin-like protein (TLP, Q5DJS5), which was identified in both the iTRAQ and YTHS analyses was the most affected being 2.3-fold up-regulation [28]. This protein is known to be an important pathogenesis-related (PR) protein belonging to the PR-1 to PR-17 family that is involved in host defense and developmental processes in plants [29,30]. Many TLP genes have been validated via empirical experiments as being associated with increased resistance to pathogen infections in transgenic plants [29]. For example it has been found that TLPs can be induced during the hypersensitive response to cucumber mosaic virus (CMV) and that they specifically interact with the CMV-MP and -CP in transgenic yeast models [31]. It is therefore interesting to note that the current study found that TLPs can also be up-regulated in cucumber plants expressing the CGMMV-MP, and those previous studies have shown that TLPs are candidate genes with the potential to create cucumber varieties resistant to CGMMV infection. The most significantly down-regulated protein (1.4-fold) in the current study was cytochrome b6-f (cyt-b6-f), which is in agreement with the iTRAQ study that found this protein was also down-regulated in response to CGMMV infection. The cyt-b6-f complex is an important protein in chloroplasts having a critical function in PS I and II and ATP synthase during photosynthesis. In addition, it has also been found to be an important component of the plant pathogen interaction, with one study finding that cyt-b6-f was inhibited in rice (Oryza sativa) plants infected by rice stripe virus (RSV) [32] causing reduced energy production and reduced synthesis of structural components of the chloroplast, which were linked to the various symptoms of infection. Furthermore, it was also found that the accumulation of RSV altered the expression of 9788 genes affecting many aspects of the host's cellular system including protein synthesis systems, organelle function, cell structure and defense systems. These studies might therefore suggest that the down-regulation of cyt-b6f could negatively affect the chloroplasts of the transgenic cucumbers and lead to reduced resistance to CGMMV.
The four other genes that had significantly altered expression in the transgenic cucumber seedlings included disulfide isomerase (PDI), cucumber peeling cupredoxin, histone H4 and cytochrome oxidase. The PDI, which was down-regulated 0.6-fold, is known to be involved in the oxidative folding of cystine knot defense proteins [33]. These results are in contrast to a previous study that found that PDI was up-regulated in Nicotiana benthamiana plants infected with Potato virus X (PVX) [34]. Furthermore, positional cloning has confirmed that variants of PDI like 5-1 (HvPDIL5-1) are linked to the Bymovirus resistance that occurs naturally in barley (Hordeum vulgare L.) [35]. Although the role of PDI is complicated, it is likely that its down-regulation in the transgenic cucumbers would have a negative effect overall, and reduce their resistance to infection. The three remaining genes that had altered expression in the transgenic cucumber seedlings were found to be up-regulated. The cucumber peeling cupredoxin, which is a common copper-binding protein, was found to be 0.8-fold up-regulated. Previous studies have shown that cupredoxin are an important factor contributing to symptoms of mottle and mosaic variegation during virus infections, which inevitably affects the photosynthesis of the host causing reduced yields [36]. It is therefore possible that the increased expression of the cucumber peeling cupredoxin in the transgenic cucumber seedlings could mitigate the symptom of CGMMV infection. Histone H4 was also found to be up-regulated in the transgenic cucumber plants. It is known that this protein can affect many developmental processes including root growth [37], flowering time [38] and seed development [39], cell wall development and plant defense response [40]. In addition, research has shown that infections of plant pathogens can lead to histone acetylation and methylation [41], and that mutations in histones can facilitate disease resistance in plants [42], which suggests that the up-regulation of histone H4 in the transgenic cucumber plants could enhance their resistance to infection. Cytochrome oxidase, which is located in the plant mitochondria and found to interact with the CGMMV-mp in the YTHS analysis, was also up-regulated in the transgenic cucumber plants. This protein has previously been shown to have a role in RNA editing and can negatively affect the viral gene silencing process [43], which indicates that cytochrome oxidase might contribute to CGMMV resistance in the GM-cucumbers.
In summary, the current study found strong evidence the introduction of transgenes into the cucumber genome has the potential to affect the expression of endogenous genes. Perhaps the most interesting effect was the down-regulation of cyt-b6-f, which indicates that the CGMMV-MP transgene has the potential to interact with the PSII of cucumber and not only increase disease resistance to CGMMV, but also suppress the expression of some resistance genes. It is well known that the introduction of foreign genes into the genome of crop plants can affect their nutritive value or alter their resistance to virus infection [44,45]. Furthermore, previous research has also demonstrated that the interaction of multiple genes in complex biological networks [46], which indicate that a wide range of factors should be assessed when considering the development of transgenic cucumbers resistant to CGMMV infection. It is also interesting to note that six of the related genes assessed in the current study were unaffected by the expression of the CGMMV-MP in the transgenic cucumber seedlings, including cysteine synthase, NADH-quinone oxidoreductase subunit K, pathogen regulatory protein CuPi1, NADH-quinone oxidoreductase subunit J, catalase and phloem protein (PP2), even though the iTRAQ and YTHS studies had suggested that they had altered expression in the response of cucumber plants to CGMMV infection. Although the current study provides important information regarding the effect of the CGMMV-MP on 12 related genes in cucumber seedlings, further research is required to characterize the effect in adult plants exposed to CGMMV and at different developmental stages to characterize the relationships between PR-genes and phenotypic changes that occur due to CGMMV infection, and also assess their genetic stability of resistance to the next generation. However, the data collected so far has provided a greater understanding of the role of pathogenesis-related proteins in transgenic cucumber seedlings, and highlighted resistance genes that have the potential to prevent CGMMV infection.
Acknowledgment
This work was supported by the National Science Foundation of China (NSFC) project (Grant No. 31371910), the Program for Changjiang Scholars and Innovative Research Team in University (Grant No. IRT1042) and the Special Fund for Agroscientific Research in the Public Interest of China (Grant No. 201303028).
References
- Ainsworth GC (1935) Mosaic diseases of the cucumber. Ann Appl Biol 22: 55-67.
- Inoue T, Inoue N, Asatani M, et al. (1967) Studies on cucumber green mottle mosaic virus in Japan. Nogaku Kenkyu 51: 175-186.
- Antignus Y, Pearlsman M, Ben-Yoseph R, et al. (1990) Occurrence of variant of cucumber green mottle mosaic virus in Israel. Phytoparasitica 18: 50-56.
- Vani S, Varma A (1993) Properties of cucumber green mottle mosaic virus isolated from water of river Jamuna. Indian Phytopathol 46: 118-122.
- Budzanivska IG, Rudneva TO, Shevchenko TP, et al. (2007) Investigation of Ukrainian isolates of cucumber green mottle mosaic virus. Arch Phytopathol Plant Protection 40: 376-380.
- Kim OK, Mizutani T, Natsuaki KT, et al. (2010) First report and the genetic variability of Cucumber green mottle mosaic virus occurring on bottle gourd in Myanmar. J Phytopathol 158: 572-575.
- Ling KS, Li R, Zhang W (2014) First report of cucumber green mottle mosaic virus infecting greenhouse cucumber in Canada. Plant Dis 98: 701.
- Tian T, Posis K, CJ Maroon-Lango, et al. (2014) First report of cucumber green mottle mosaic virus on melon in the United States. Plant Dis 98: 1163.
- Tesoriero LA, Chambers G, Srivastava M, et al. (2015) First report of cucumber green mottle mosaic virus in Australia. Australasian plant dis. Notes 11: 1.
- Hollings M, Komuro Y, Tochihara H (1975) Cucumber green mottle mosaic virus. Descriptions of plant Viruses 154.
- Choi GS (2001) Occurrence of two tobamovirus diseases in cucurbits and control measures in Korea. Plant Pathol J 17: 243-248.
- Baker C (2013) Cucumber green mottle mosaic virus (CGMMV) found in the United States (California) in melon. Pest alert, Florida department of agriculture and consumer services, division of plant industry DACS-P-01863.
- De-Both MTJ, Fierens OEV (2004) Methods for generating resistance against CGMMV in plants. United States Patent Application Publ No. US 2004/0237136 A1.
- Park SM, Lee JS, Jegal S, et al. (2005) Transgenic watermelon rootstock resistant to CGMMV (cucumber green mottle mosaic virus) infection. Plant Cell Rep 24: 350-356.
- Taliansky M, Torrance L, Kalinina NO (2008) Role of plant virus movement proteins. Methods Mol Biol 451: 33-54.
- Vogler H, Kwon MO, Dang V, et al. (2008) Tobacco mosaic virus movement protein enhances the spread of RNA silencing. PLoS Pathog 4: e1000038.
- Huppert E, Szilassy D, Salánki K, et al. (2001) Heterologous movement protein strongly modifies the infection phenotype of cucumber mosaic virus. J Virol 76: 3554-3557.
- Zeller SL, Kalinina O, Brunner S, et al. (2010) Transgene × Environment interactions in genetically modified wheat. PLoS ONE 5: e11405.
- Herbers K, Tacke E, Hazirezaei M, et al. (1997) Expression of a luteoviral movement protein in transgenic plants leads to carbohydrate accumulation and reduced photosynthetic capacity in source leaves. Plant J 12: 1045-1056.
- Heuberger S, Crowder DW, Brvault T, et al. (2011) Modeling the effects of plant-to plant gene flow, larval behavior, and refuge size on pest resistance to Bt cotton. Environ Entomol 40: 484-495.
- Sarkar B, Patra AK, Purakayastha TJ (2008) Transgenic Bt-cotton affects enzyme activity and nutrient availability in a sub-tropical inceptisol. J Agron Crop Sci 194: 289-296.
- De-Francesco L (2013) How safe does transgenic food need to be? Nature Biotechnol 31: 794-802.
- Beachy RN (1995) Transgenic virus-resistant plants and new plant virus, movement protein-Mediated resistance. The USDA's Animal and Plant Health Inspection Service (APHIS) and the America Institute of Biological Sciences (AIBS).
- Liu HW, Luo LX, Li JQ, et al. (2014) Pollen and seed transmission of cucumber green mottle mosaic virus in cucumber. Plant Pathol 63: 72-77.
- Antignus Y, Lapidot M, Ganaim N, et al. (1997) Biological and molecular characterization of tomato spotted wilt virus in Israel. Phytoparasitica 25: 319-330.
- Cheng YQ (2008) Studies on the molecular interactions between SCMV HC-Pro and maize proteins. China Agricultural University, Beijing, China.
- Guo JY, Song LN, Ma SC, (2004) Rapid LacZ screening method increases the sensitivity of yeast two-hybrid screening. Biotechnol 14: 31-33.
- Liu HW, Liang CQ, Liu PF, et al. (2015) Quantitative proteomics identifies 38 proteins that are differentially expressed in cucumber in response to cucumber green mottle mosaic virus infection. Virol J 12: 216.
- Liu JJ, Sturrock R, Ekramoddoullah AKM (2010) The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rpt 29: 419-436.
- Petre B, Major I, Rouhier N, et al. (2011) Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol 11: 33.
- Kim MJ, Ham BK, Kim HR, et al. (2005) In vitro and in planta interaction evidence between Nicotiana tabacum thaumatin-like protein 1 (TLP1) and cucumber mosaic virus proteins. Plant Mol Biolol 59: 981-994.
- Satoh K, Kondoh H, Sasaya T, et al. (2010) Selective modification of rice (Oryza sativa) gene expression by rice stripe virus infection. J Gen Virol 91: 294-305.
- Gruber CW, Cemazar M, Clark RJ, et al. (2007) A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J Biol Chem 282: 20435-20446.
- Ye C, Dickman MB, Whitham SA, et al. (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156: 741-755.
- Yang P, Lüpken T, Habekuss A, et al. (2014) Protein disulfide isomerase like 5-1 is a susceptibility factor to plant viruses. Proc Natl Acad Sci 111: 2104-2109.
- Choi M, Davidson VL (2011) Cupredoxins-a study of how proteins may evolve to use metals for bioenergetic processes. Metallomics 3: 140-151.
- Yao X, Feng H, Yu Y, et al. (2013) SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLoS ONE 8: e56537.
- Cao Y, Dai Y, Cui S, et al. (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586-2602.
- Wang Z, Cao H, Chen FY, et al. (2014) The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem 84: 125-133.
- Rosa S, Ntoukakis V, Ohmido N, et al. (2014) Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell 26: 4821-4833.
- De-La-Peña C, Rangel-Cano A, Alvarez-Venegas R (2012) Regulation of disease-responsive genes mediated by epigenetic factors: interaction of Arabidopsis-Pseudomonas. Mol Plant Pathol 13: 388-398.
- Choi SM, Song HR, Han SK, et al. (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J 71: 135-146.
- Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49: 311-343.
- Kanobe MN, Rodermel SR, Bailey T, et al. (2013) Changes in endogenous gene transcript and protein levels in maize plants expressing the soybean ferritin transgene. Front Plant Sci 4: 196.
- Fitchen JH. Beachy RN (1993) Genetically engineered protection against viruses in transgenic plants. Annu Rev Microbiol 47: 739-763.
- Hu JK, Wang X, Wang P (2014) Testing gene-gene interactions in genome wide association studies. Genet Epidemiol 38: 123-134.
Corresponding Author
Jianqiang Li, Ph.D, Department of Plant Pathology, China Agricultural University, 100193 Beijing, P.R. China, Tel: 86-10-62734938.
Copyright
© 2016 Liu H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.