Hypothetical Origin of NAT2 Slow Acetylator in Human Evolution
Abstract
In the process of biological evolution, our ancestors have faced many environmental challenges and the genomes of the present-day humans are storages of environmentally adapted genetic entities in the form of functional genes. To this respect, NAT2 a drug metabolizing gene plays a vital role in differential metabolization rate due to genomic changes that result in changed amino acids. Different alleles in genomes are well correlated with differential acetylation mechanism leading to inter-individual variation with respect to drug response. This correlation prompted us to determine the mutational trajectory of slow acetylator NAT2 alleles in Indian population that are responsible for low Nat2 enzymatic activity. With the overwhelming increase in published NAT2 coding data (873 bp), characterization of acetylator phenotypes were based on the seven SNP panel as described in several studies. Among the slow acetylator alleles, NAT2*5B, NAT2*6A and NAT2*7B were found to be highly prevalent in Indian population. Out of these, NAT2*5 alleles were highly mutable as compared to the NAT2*6A and NAT2*7B that substantiates the hypothesis that neolithic transition has contributed the emergence of vast majority of mutations due to exposure of exogenous chemicals.
Keywords
Genomes, Mutational, Acetylation, Neolithic
Introduction
The human N-acetyltransferase 2 is a broad-spectrum drug metabolizing gene that plays a role in the metabolization of a wide variety of toxins and clinically important drugs, like anti-tuberculosis (Isoniazid), anti-malarial (Dapsone/Sulphadoxine) and etc [1]. NAT2 gene, which exhibits a well-established acetylation polymorphism, categorized individuals into fast and slow acetylators and has been the focus of numerous studies due to ongoing targets of population-specific selective pressures which is stimulated by the patterns of geographic differentiation of major alleles [2]. For example, acetylation profiling of ethnically diverse populations displayed role of natural selection in shaping genetic variation at this locus based on NAT2 DNA sequence [3-7]. It has been well documented that Dietary evolution is fundamental to human history [8] and people lived as hunter-gatherers using range of plants and animals in their natural environment. Around 10,000-12,000 years ago, the process of Neolithisation started near Fertile crescent region with the beginning of agriculture and animal husbandry that resulted in the most radical change in human economic and subsistence strategies [9]. Food resources became more and more abundant and the consumption of cereals and other plants increased tremendously [10]. The domestication of plants and animals led to changes in numerous aspects of life, including available foodstuffs, physical activities, reproductive experience, psychosocial relations, microbial interactions, toxin/allergen exposure, and sedentism. This Neolithisation has made a powerful impact on the human genome which was also demonstrated in certain genes such as LCT, AMY and PLRP2, influencing differential patterns of gene expression among human population [11-13]. Furthermore, NAT2 gene plays a vital role in mediating interaction between xenobiotics and humans that varies and are dependent on diet (component of environment) making them excellent target for the action of natural selection. Several studies have advocated the positive correlation between different acetylator phenotype (fast and slow) and differential subsistence mode strategies (hunter gatherers and agriculture/pastoralist) [3,4,6]. Several genetic surveys have provided clear evidence, correlating different acetylator phenotypes with different subsistence mode of strategies (adopted by their ancestors) in the last 10,000 years, suggesting a possible selection in populations shifting from hunting to farming [3,7]. Thus, it is hypothesized that the neolithic transition has triggered significant changes in dietary exposure that modified the substrate specificity affecting the Nat2 acetylation pathway. Interestingly, with the availability of archaic genome dataset [14], it was found that archaic hominin harbors fast acetylator phenotype which indicates that effect of neolithization on modern human population and emergence of slow acetylator.
Considering the fluctuations in the type and levels of xenobiotics compounds over time immemorial, the metabolic function of Nat2 is likely to be under adaptive evolution. Owing to this, NAT2 gene could broaden or change its substrate specificity in response to change in environmental conditions. Despite the wide characterization of NAT2 gene in global human population with respect to population genetic studies and association studies (NAT2 gene and diseases), no study to date has investigated the mutational trajectory of the non-synonymous allele which affects the activity of NAT2. The aim of the present work is to thoroughly examine the mutational trajectory of three diverse slow NAT2 acetylator haplotypes (NAT2*5, NAT2*6, NAT2*7) which are common in various human population, utilizing SNPs in NAT2 coding region.
Materials and Method
Selection of region
Genetic basis of acetylation process is determined by genotypic characterization of the one coding exon (873 bp) NAT2 gene, located on the short arm of human chromosome 8 [2,3,7] Based on the SNPs present in the NAT2 coding region, the data of NAT2 SNPs were retrieved from NAT2 nomenclature website (http://nat.mbg.duth.gr/) and the mutational trajectories of slow acetylators NAT2*5, NAT2*6 , NAT2*7 were tracked.
Results
NAT2 alleles, acetylation phenotype and mutational trajectories of non-synonymous mutations
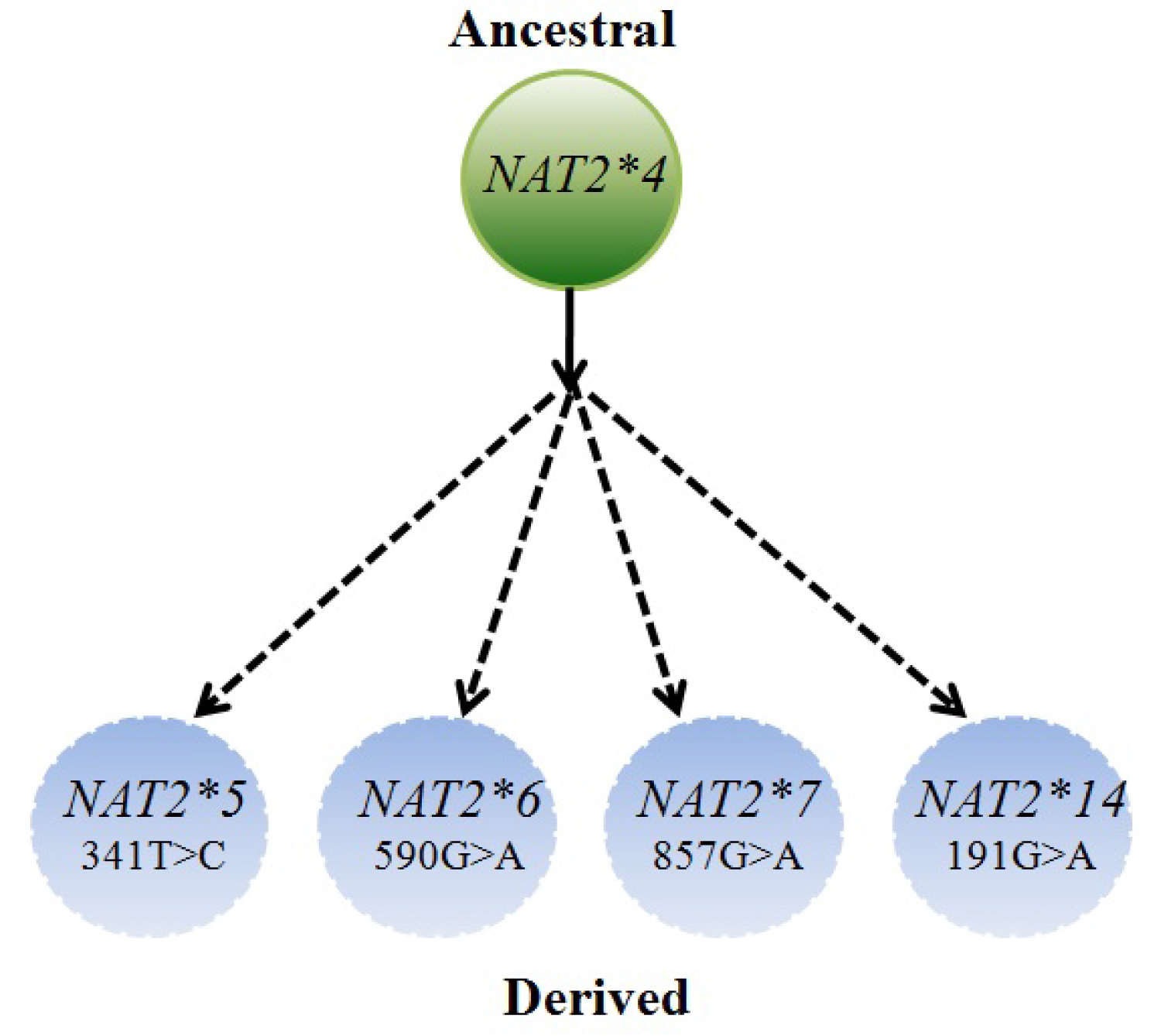
With the availability of NAT2 data (NAT2 nomenclature), NAT2 alleles were predicted based on seven SNPs reported in coding region (191 G > A, 282C > T, 341T > C, 481T > C, 590G > A, 803A > G, 857G > A). Out of these, five (191, 341, 590, 803 and 857) were non-synonymous mutation leading to significant decrease in acetylation capacity, whereas two (282, 481) were found to be synonymous that does not alter protein activity. Based on the combination of these SNPs, 20 haplotypes have been identified so far (http://nat.mbg.duth.gr/). Out of these haplotypes, NAT2*4, a reference allele is an ancestral and a predictor of fast acetylator phenotype. Moreover, NAT2*5, NAT2*6 and NAT2*7 haplotypes were diverse and predictor of slow acetylator. NAT2*5, NAT2*6 were found to be most common haplotypes, whereas NAT2*7 allele was found to be relatively less among population (Figure 1). To be noted, present study is focused on haplotypes that are predictors of slow acetylators, hence the fast acetylators haplotypes was not included. In addition, 191 G > A that forms NAT2*14 haplotype was restricted to African population, and thus not considered in the present study analysis.
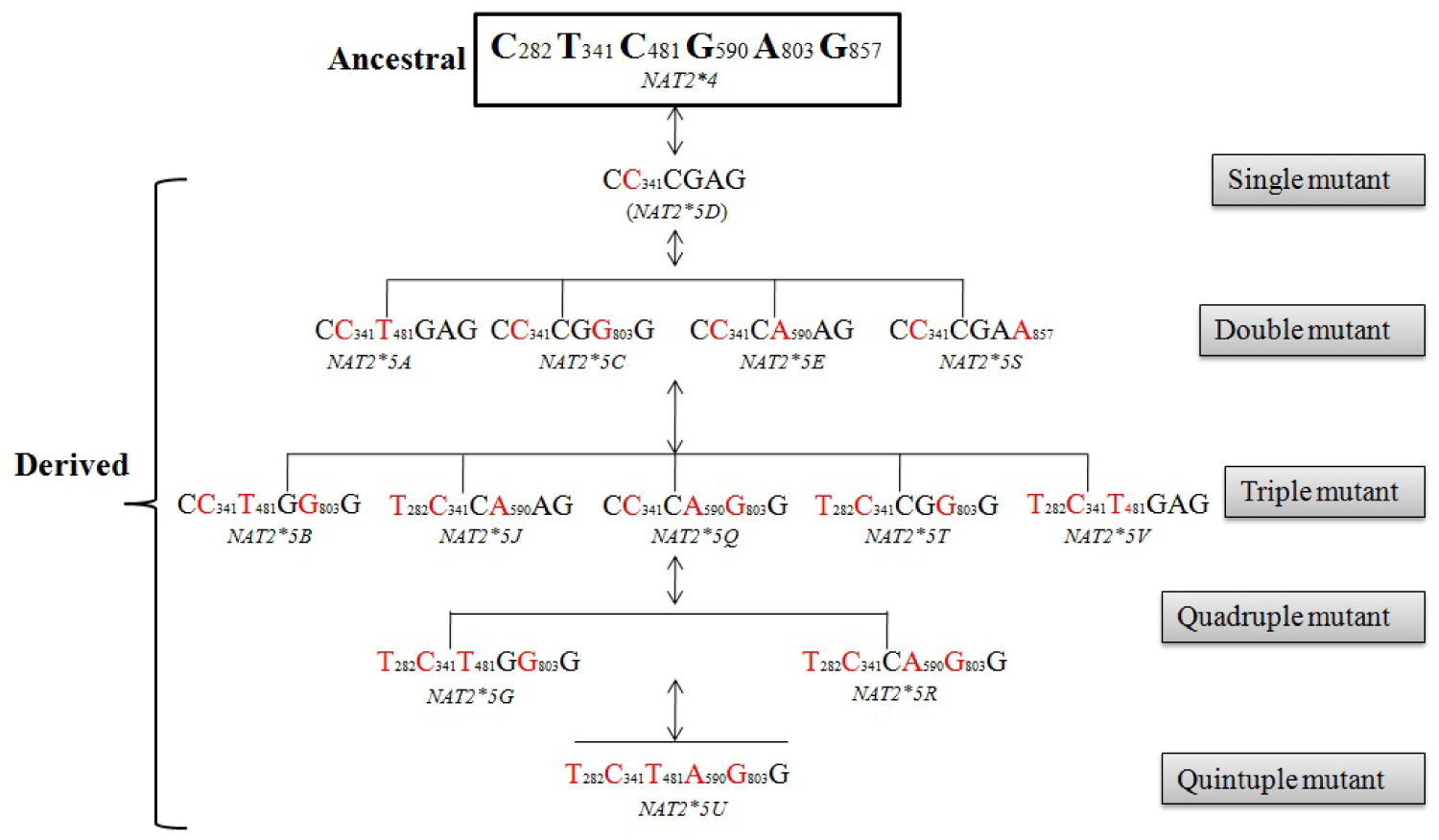
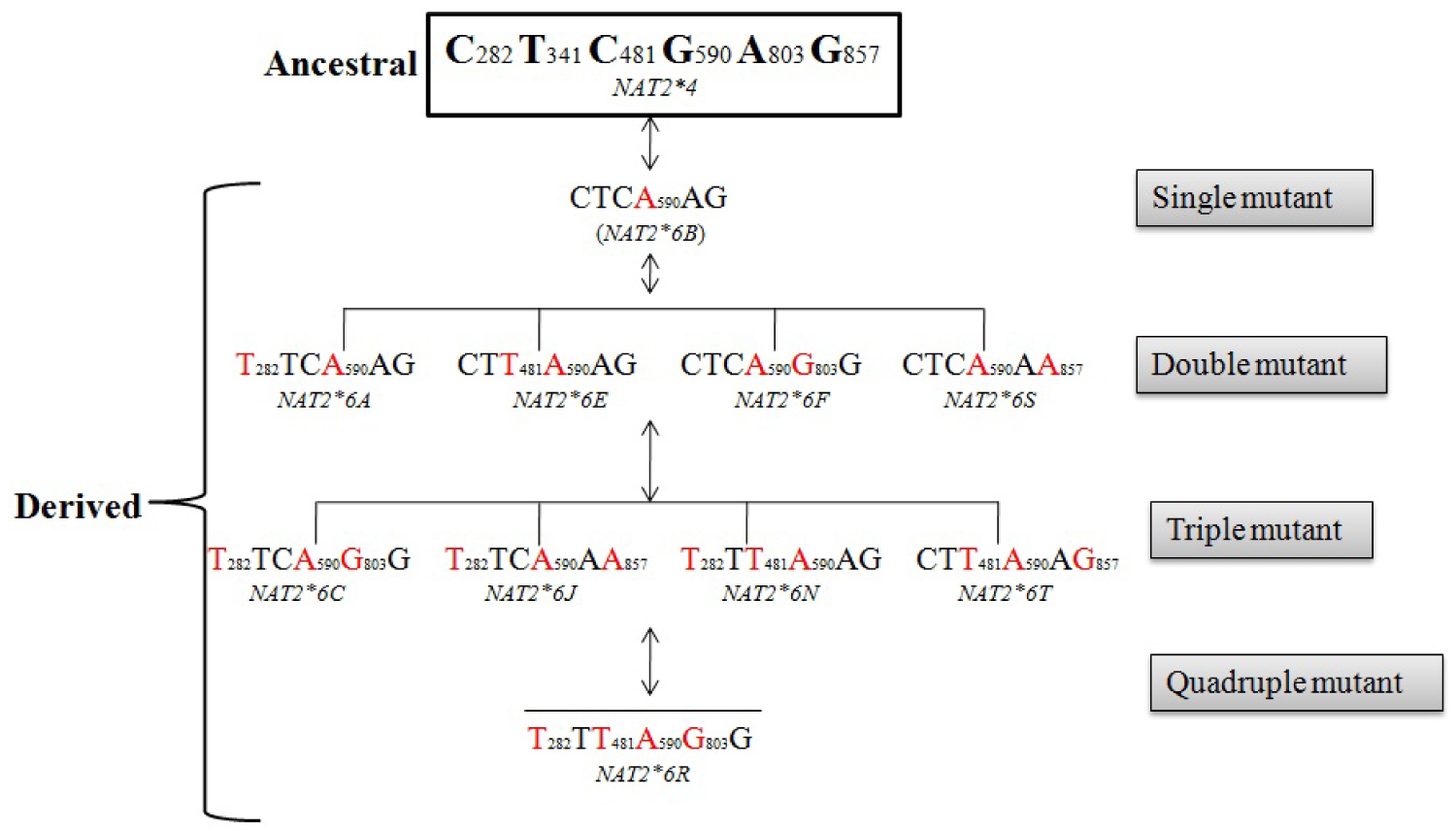
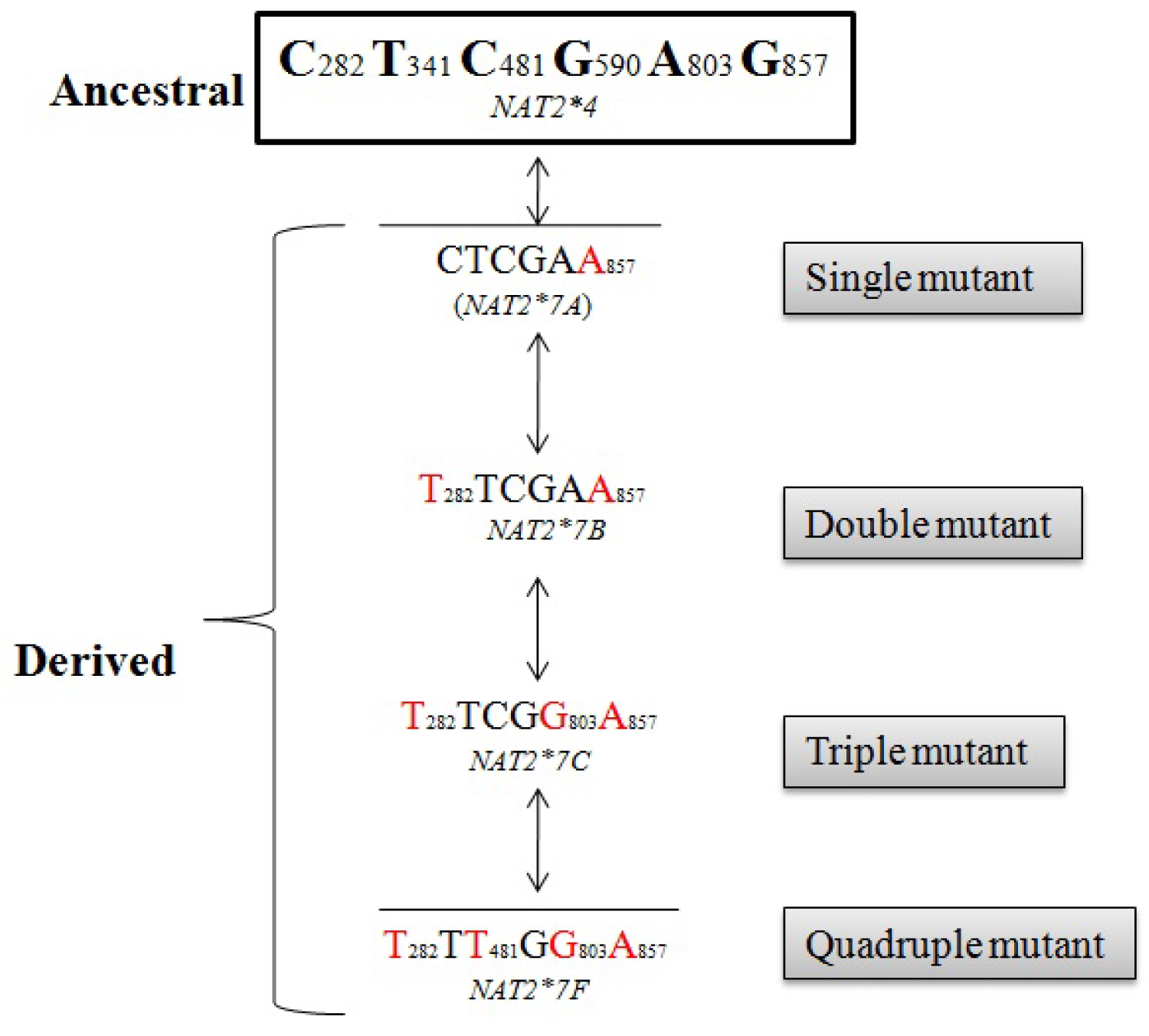
Taken into consideration the three common NAT2 haplotypes, we herewith, infer possible mutational trajectories from the ancestral NAT2*4 to derived NAT2 slow haplotype (NAT2*5, NAT2*6 and NAT2*7). A hypothetical sketch of the possible mutational events from ancestral to derived alleles is shown in Figure 2, Figure 3 and Figure 4. It is evident from the Figure 2, that NAT2*5 mutational trajectory shows highest number of double (4), triple (5) and quadruple (2) mutants. Also, it contains quintuple mutant which is formed by the combination of five mutations. while, NAT2*6 harbors double (4), triple (4) and quadruple (1) mutant (Figure 3), NAT2*7 shows least number of mutants with double (1), triple (1) and quadruple (1) mutant (Figure 4).
Discussion
With the advancement of high throughput DNA technologies and human genome research (high coverage), new tools are utilized to identify variations in both mitochondrial as well as nuclear DNA [15-18]. These variations help in understanding the differences between human and other mammalian species (including our evolutionary closest relatives Great Apes) and constitute a key driving force in evolution [19,20]. They are the main sources of evolutionary novelty that acts on the genetic constitution of an organism at different levels, from single nucleotide substitutions to large-scale chromosomal modifications [1]. Genetic analysis of mutations allowed us to estimate the strength of natural selection that favors organisms, best adapted to their respective surroundings, and shed light on episodes of adaptive evolution at specific sites and domains of the protein. Understanding the mutational pathway of non-synonymous polymorphisms would help in drug targets and predict the effects of mutations on diseases. It is thus essential to analyze the mutational trajectories of the non-synonymous polymorphisms that alter the activity of protein. To this respect, NAT2 gene is widely considered as the potential target of selective pressure varies across different populations that can be related to dietary habits and environmental conditions influencing Nat2 expression. It was documented that the slow acetylators were originated with the advent of agriculture 10,000 years ago near Fertile Crescent region [3,6]. This neolithic transition has made a powerful impact on the human genome influencing differential patterns of gene expression [11-13]. Interestingly, the archaic hominin genome (Altai and Vindijia Neanderthal) showed only synonymous variants (C282T, and G803A; indicator of fast acetylator phenotype) [14,21], further supports our hypothesis that slow acetylators emerged as a result of Neolithic transition.
Among the derived alleles (slow acetylator phenotype), it was reported earlier that the NAT2*5 was common in Western and Central Eurasians, NAT2*6 was found around the globe in intermediate frequency and NAT2*7 was observed mainly in Eastern Eurasians. Considering three NAT2 slow haplotype (NAT2*5, NAT2*6, and NAT2*7), NAT2*5 harbors maximum number (single quintuple) of mutants as compared to NAT2*6 and NAT2*7. It is also interesting to note that among the slow acetylator haplotype NAT2*5, NAT2*5B was found in high frequency which further strengthen the hypothesis that, NAT2*5B allele might be originated with the advent of agriculture during neolithic transition as a consequence of exposure to exogenous chemicals [3]. Similarly, among the other slow acetylator haplotype NAT2*6 and NAT2*7 , NAT2*6 A and NAT2*7 B was found in highest frequency which was thought to be earlier present in two agriculturalist population (Tajik and Kazakh) of Central Asia [6].
In conclusion, among NAT2 slow acetylator haplotypes, NAT2*5 harbors highest mutant and hypothesized to be originated with the transition of subsistence mode from hunting to agriculture during human evolution. Furthermore, these mutational trajectories would help us in understanding the role of mutation occurrence which can be further associated with the susceptibility of diseases and adverse drug reactions.
Conflict of Interest
There is no conflict of interest.
References
- Khan N, Chand A (2015) Antimalarial metabolization: What have we learnt so far? World Journal of Pharmacy and Pharmacology 1: 002-010.
- Khan N, Pande V, Das A (2011) Characterization, comparative genomics, and evolutionary inferences of a human drug metabolizing (NAT2) gene. Interventional Medicine and Applied Science 3: 65-73.
- Patin E, Barreiro LB, Sabeti PC, et al. (2006) Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes. Am J Hum Genet 78: 423-436.
- Fuselli S, Gilman RH, Chanock SJ, et al. (2007) Analysis of nucleotide diversity of NAT2 coding region reveals homogeneity across Native American populations and high intra-population diversity. Pharmacogenomics J 7: 144-152.
- Luca F, Bubba G, Basile M, et al. (2008) Multiple advantageous amino acid variants in the NAT2 gene in human populations. PLoS One 3: e3136.
- Magalon H, Patin E, Austerlitz F, et al. (2008) Population genetic diversity of the NAT2 gene supports a role of acetylation in human adaptation to farming in Central Asia. Eur J Hum Genet 16: 243-251.
- Khan N, Pande V, Das A (2013) NAT2 sequence polymorphisms and acetylation profiles in Indians. Pharmacogenomics 14: 289-303.
- Hublin JJ, Richards MP (2009) The evolution of hominin diets. Integrating approaches to the study of palaeolithic subsistence. Springer-Verlag, Dordrecht, Netherlands.
- Eaton SB, Cordain L (1997) Evolutionary aspects of diet: Old genes, new fuels. Nutritional changes since agriculture. World Rev Nutr Diet 81: 26-37.
- Simmons AH (2007) The Neolithic Revolution in the Near East. Transforming the human landscape. University of Arizona Press, Tucson.
- Coyne JA, Hoekstra HE (2007) Evolution of protein expression: New genes for a new diet. Current Biology 17: R1014-R1016.
- Eisenstein M (2010) Diversity: Of beans and genes. Nature 468: S13-S15.
- Hancock AM, Alkorta-Aranburu G, Witonsky DB, et al. (2010) Adaptations to new environments in humans: The role of subtle allele frequency shifts. Philos Trans R Soc Lond B Biol Sci 365: 2459-2468.
- Prüfer K, Filippo CD, Grote S, et al. (2017) A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358: 655-665.
- Khan N, Chittoria C, Pande V, et al. (2012) Development of multilocus putatively neutral DNA markers in the X-chromosome for population genetic studies in humans. Ann Hum Biol 39: 281-289.
- Bhatt N, Khan N, Dash SK, et al. (2014) nspA Gene as a specific genetic marker for detection of Neisseria meningitidis causing bacterial meningitis. Indian J Biochem Biophys 51: 211-214.
- Khan N (2016) Recent advancements in diagnostic tools in mitochondrial energy metabolism diseases. Advances in Medical sciences. 61: 244-248.
- Khan N (2015) Comparative and evolutionary insights into CD4 gene across mammalian and avian taxa. Interventional Medicine and Applied Science 7: 152-160.
- Khan N, Kim SK, Gagneux P, et al. (2019) Maximum reproductive lifespan correlates with CD33rSIGLEC gene number: Implications for NADPH oxidase-derived reactive oxygen species in aging. The FASEB Journal 34: 1928-1938.
- Khan N, De Manuel M, Peyregne S, et al. (2020) Multiple Genomic Events Altering Hominin SIGLEC Biology and Innate Immunity Predated the Common Ancestor of Humans and Archaic Hominins. Genome Biology and Evolution 12: 1040-1050.
- Prüfer K, Racimo F, Patterson N, et al. (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43-49.
Corresponding Author
Dr. Ratan Kumar Rai, University of Kentucky, Lexington, Kentucky, USA.
Copyright
© 2020 Rai RK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.