Characterization of Drought Responsive Genes of CIPK Families in Rice, Maize and Sorghum
Abstract
The CIPK gene family plays a key role in plant development and in stress signal transduction. The orthologs for experimentally proven drought stress responsive CIPKs from rice was identified in maize and sorghum. A total of 49 genes from the three species were analyzed for their phylogenetic relationship, gene structure, expression level and tissue specificity upon drought stress. The drought stress tolerance specificity of intronless group of CIPKs and multi-stress responsive nature of intron rich CIPKs were identified. The group level characterization revealed the similarity in function among Group I and Group II (A) CIPKs as the number and distribution of motifs were similar. Functional similarity of the genes was analyzed by in-silico expression analysis using publicly available data and which confirmed the drought responsiveness of 31 genes as they had similar expression level and it also shows the conservation of functions between species.
Keywords
CIPK, Drought, Stress, Intron, Protein Kinase
Introduction
Climate change induced by emission of green house gas can cause severe drought [1,2]. The higher plants adopt numerous mechanisms to cope with drought. The role of protein kinase gene family in drought stress response was revealed by many genome-wide gene expression profiling studies and pointed that the drought stress response given by them are efficient, fast-acting and reversible [3-5] responses to many environmental stresses such as salinity, cold and drought [6]. The changes in concentration levels of Ca2+ are recognized by several Ca2+ binding proteins including calmodulin (CaM), calmodulin like proteins (CMLs), Ca2+-dependent protein kinases (CDPKs) and calcineurin B-like proteins (CBLs) and results in downstream responses [7,8]. CDPKs are exceptional in this category as they have a kinase domain and other three Ca2+ sensors had no enzymatic domains. Except CDPKs, other Ca2+ sensors interact with their respective target proteins and modulate their activity [8]. Whereas, CDPKs serves as special sensor as they directly initiate the downstream phosphorylation events up on Ca2+ binding due to the presence of CaM like and protein kinase domains [9]. The target protein of CBLs is referred to as CBL-interaction protein kinases (CIPKs) [7] and is also known as SnRK3. CIPK proteins consist of a conserved N-terminal kinase domain followed by junction domain and C-terminal regulatory domain. The Ca2+ bound CBL proteins interact with target protein CIPK through a conserved NAF/FISL motif at the C terminal regulatory domain of CIPK and activate its catalytic activity [10]. Total of 33 CIPKs was identified in rice through bioinformatics analysis [11,12], 43 CIPKs are identified in maize [13] and 32 CIPKs are identified in sorghum [14]. CIPKs are reported to be expressed in response to various stresses. Over expression of OsCIPK23 improved drought tolerance in rice [15]. In Arabidopsis AtCIPK24 and AtCIPK7 contribute to salt and cold stress [16,17]. A cotton CIPK gene GhCIPK6 was over expressed in Arabidopsis and found that the tolerance of the plant increased in drought stress [18].
The present study intended to expose the characteristics of potential drought responsive ortholog genes in maize and sorghum by comparative analysis of them with the experimentally proven drought responsive genes of rice. The drought responsive rice genes of CIPKs (CIPK1, CIPK2, CIPK5, CIPK9, CIPK11, CIPK12, CIPK15, CIPK17, CIPK20, CIPK21, CIPK22, CIPK23, CIPK24, CIPK29 and CIPK 30) [19] were selected for the study as they have experimental evidences.
Materials and Methods
Datasets
Genomic, CDS and protein sequences of rice genes were retrieved from Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). Ensembl Plants (http://plants.ensembl.org) and NCBI (https://www.ncbi.nlm.nih.gov) were used for retrieving maize and sorghum gene sequences. The retrieved protein sequences of rice were subjected to a BLASTp (https://blast.ncbi.nlm.nih.gov) analysis to find the orthologous genes in maize and sorghum by means of reciprocal best hit approach. The protein sequences which showed identity ≥ 75% were considered as orthologous. The orthologous genes for CIPK 20, CIPK 22, CIPK 29 and CIPK 30 were not identified in sorghum and maize. Therefore a total of 49 protein sequences were analyzed further which included the homologues sequences of rice in sorghum and maize.
Phylogenetic analysis
The multiple sequence alignment of 49 full length protein sequences of all the three species were constructed using CLUSTALW [20]. Phylogenetic tree was constructed by using Neighbor-Joining method by considering 1,000 rapid bootstrap replicates with the help of MEGA X [21] and it was visualized using iTOL (http://itol.embl.de). The aligned sequence file was also used for finding out the discrete Gamma distribution to recognize the evolutionary rate difference. Number of discrete categories used for the analysis was 5. Substitution pattern and rates were estimated under the Jones-Taylor-Thornton [22] model (+G) [22]. The tree topology was automatically computed in MEGA X for estimating ML values.
Characterization of phylogenetic groups
The gene structures of all the genes were predicted by aligning the coding sequence with its corresponding genomic sequence by using GSDS 2.0 server (http://gsds.cbi.pku.edu.cn). GSDS 2.0 is an improved version of GSDS and it supports two more widely used annotation formats, providing more comprehensive support for annotation files. To identify the conserved motifs in each group, the complete sequence of proteins at the groups were submitted to MEME suite (http://meme.sdsc.edu/meme/) [23]. This tool discovers the ungapped motifs in the sequence and splits the variable-length patterns in to more than two unique motifs. For the analysis we used the optimum width of motifs ranging from 12 to 60 by setting the search for 5 best motifs. The identified motifs were annotated by using Motif Scan (https://myhits.isb-sib.ch/cgi-bin/motif_scan) and InterProScan (https://www.ebi.ac.uk/interpro/search/sequence-search) [24]. The conserved domains were identified by Pfam [25]. Physiochemical properties of proteins were analyzed by using ProtParam tool available on ExPasy proteomics server (http://web.expasy.org/compute_pi) [25].
Gene expression analysis
To understand the function of the genes, expression pattern under drought stress was analyzed using GENEVESTIGATOR [27], which has a manually curated and well annotated database of expression data collected from variety of public repositories including Gene Expression Omnibus [28] and Array Express [29] (https://www.ebi.ac.uk/arrayexpress/). The gene expression at various development stages were observed for all the species. For rice, Affymetrix Rice Genome Array platform was used with water deficit microarray datasets GSE6901, GSE14275, GSE26280, E-MEXP-2401, GSE25176, GSE23211, GSE31077, GSE42683, GSE81253, GSE41647, GSE80246, GSE83378, and GSE57154. For maize, Affymetrix Maize Genome Array and mRNA- Seq Gene Level Zea Mays (ref: AGPV4) platforms were used with water deficit microarray datasets GSE16567, GSE43088 and GSE59533. In the case of sorghum the selected genes were analyzed for expression at various growing stage by using the Affymetrix Whole- transcriptome Sorghum Array platform with dataset GSE49879. The drought stress expression profile was analyzed using dataset GSE80699. The seven development stages in rice were considered for the analysis and they were seedling, tillering, stem elongation, booting, heading, flowering and milk stages. Three stages were considered in maize viz., seedling, stem elongation and anthesis. Five stages were included in sorghum analysis viz., seedling, stem elongation, booting, flowering and dough stages.
Results and Discussion
Analysis of protein properties
The nature of the proteins was analyzed by using physiochemical properties such as isoelectric point (pI), molecular weight, instability index and grand average of hydropathy (GRAVY). The pI value of the proteins ranged from 5.32 to 9.53 (Table 1). It shows that CIPK proteins have heterogeneous nature. Molecular weight of the proteins ranged from 40347.82 to 110358.1 Da. The variation in molecular weight among the members of the same group exists due to the variable number of domains contributing to protein size difference [30]. Except A0A1D6N844 all the other proteins exhibited hydrophilic nature since they had a negative GRAVY score [31]. Instability index showed that 26.5% proteins were unstable and 73.5% proteins were stable.
Phylogenetic analysis
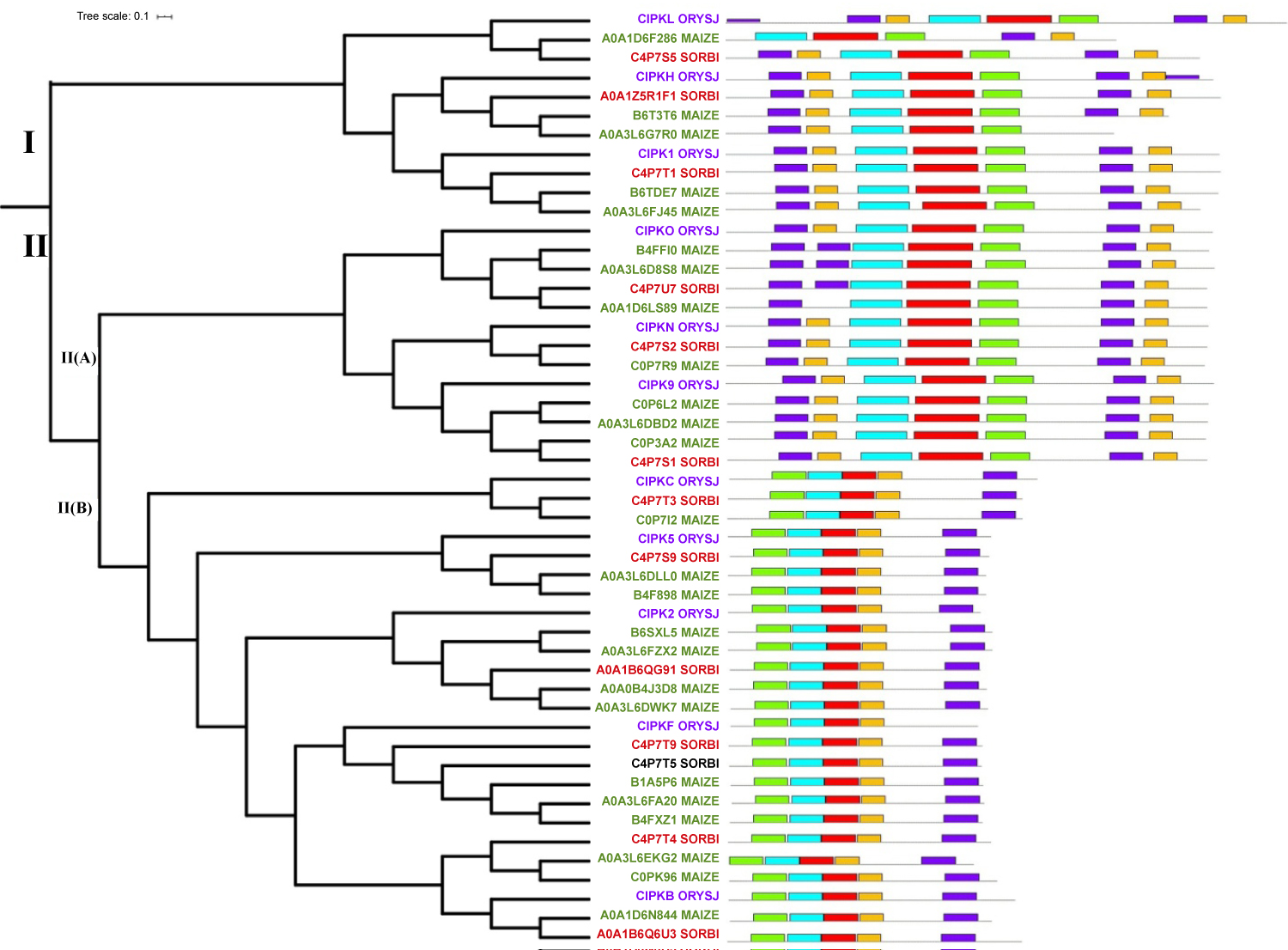
To understand the evolutionary relationship of the drought responsive genes of CIPK gene family a rooted phylogenetic tree was constructed (Figure 1). Two major groups were identified from the analysis and the Group II had subdivision. Group I consisted of 3 members of rice, 5 members of maize and 3 members of sorghum. This group was found to be the smallest group. Group II (A) consisted of 3 members of rice, 7 members of maize and 3 members of sorghum. The largest group was Group II (B) which consisted total of 25 members; 5 from rice, 13 from maize and 7 from sorghum. The estimated value of the shape parameter for the discrete Gamma Distribution was 0.7910. Total of 5 categories were considered in the analysis of sites. Mean evolutionary rates in these categories were 0.07, 0.29, 0.63, 1.20, 2.81 substitutions per site. As the shape parameter is small most of the sites evolved very slowly in the evolutionary tree [32].
Characterization of the Groups by gene structure analysis, motif analysis and protein properties analysis
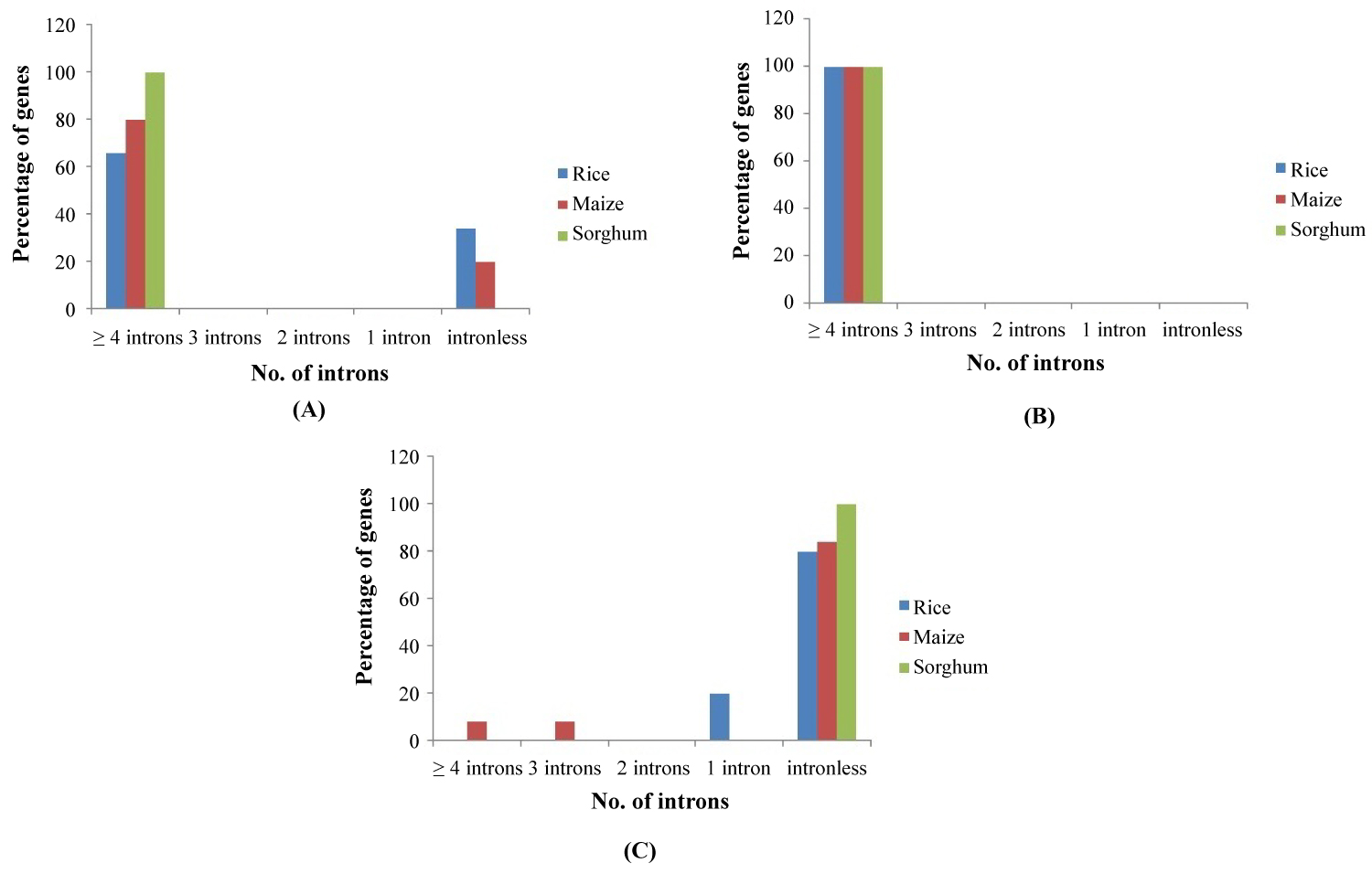
The characterization of the phylogenetic groups was carried out by gene structure analysis which showed the divergence among the groups. Above 60% of the genes in Group I had ≥ 4 introns. All the sorghum genes belonged to this category (Figure 2). In rice and sorghum 40% and 20% genes were intronless respectively. All the genes in Group II (A) showed intron richness (≥ 4 introns). The Group II (B) dominated in intronless feature. In this category all the sorghum genes were intronless. In the case of rice, 80% of genes were intronless and 20% of genes belonged to the category of genes with single intron. Similarly, 84% of genes in maize were intronless and an equal distribution of genes (8% in each category) was present in ≥ 4 introns and 3 introns category. The loss or gain of introns could have played in grouping of CIPK genes. The variation in the number of introns among the three Groups points to the genome evolution by means of selection pressure and population size [33,34]. Moreover the intron rich behavior of the genes will add to the functional diversity through alternate splicing and exon shuffling [35]. Therefore the intron rich Group I and II (A) genes might be involved in multiple pathways in response to various abiotic stress signals. The intronless feature of Group II (B) CIPK genes strongly indicates that they are single stress inducible genes and possibly the stress is drought [36]. The significance of intron poor genes in drought tolerance was analyzed by Zhu, et al. [36], and proved its role in soybean CIPK-intron poor clade genes.
The conserved motifs and their organization were identified in each group (Figure 1). It was found that all the motifs were part of protein kinase domain. The Protein kinase domain which is conserved at the N-terminus included a protein kinase ATP binding site followed by serine/threonine protein kinase active site. Group I and Group II (A) had similarity in distribution of motifs which points to their similarity in function. In Group I number of motifs varied from 5 to 8 and in Group II (A) it was varied from 6-7. Meanwhile, in Group II (B) 5 motifs were reported and all of them were highly conserved among the members. Motif 1 was either present nearby N-terminus or in the N-terminus and it represented a protein kinase ATP binding site domain. These domains are glycine rich with lysine residue in vicinity and are located at N terminus [37].
Functional analysis by gene expression level
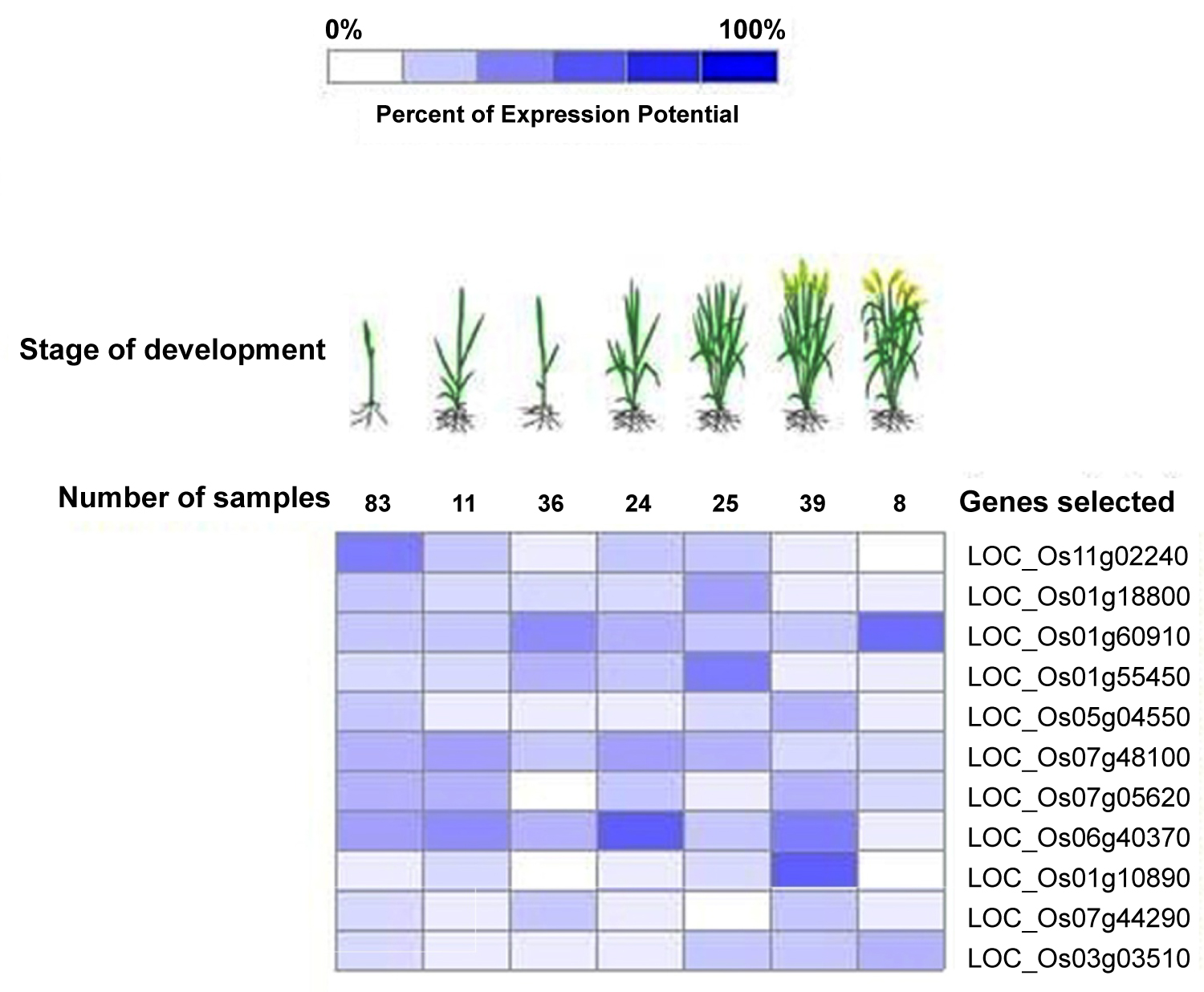
The CIPK genes play an important role in the growth and development of the plants [12,38]. Hence the expression levels at various development stages were analyzed. In rice, CIPK genes showed up regulated expression at all the stages except 4 genes, LOC_Os11g02240, LOC_Os07g05620, LOC_Os01g10890 and LOC_Os07g44290 at milk, stem elongation, milk and stem elongation and heading stages respectively as seen in Figure 3. It is also noted that they possessed high expression potential at other stages. The expression level in tissues of leaf, seedling, sheath, panicle, roots, pistil, caryopsis, flag leaf and anther were analyzed for all the genes and it was observed up regulated expression for 4 genes viz., LOC-Os11g02240, LOC-Os01g18800, LOC-Os07g48100 and LOC-Os06g40370 respectively in all the selected tissues as depicted in Table 2. Medium level of expression in all the tissues were reported for CIPK12 (LOC-Os01g55450).
In maize, the gene expression analysis was carried out by observing the expression level at three development stages with respect to drought stress. All the 9 selected genes had up regulated expression at three stages of development. The tissues analyzed for expression level were foliar leaf, shoot and root. It was observed that genes Zm00001d015325 and Zm00001d036879 had medium level of expression in all the three tissues. The gene Zm00001d000407 had low level expression in roots and medium expression in foliar leaf and shoot.
The expression of 11 genes in sorghum was analyzed at various tissues and it was observed that the gene SORBI_3003G024400 was not expressed at dough stage. All the other genes exhibited potential for expression at all development stages. The tissues analyzed for expression were rind, internode, shoot, pith, leaf and roots. The genes SORBI_3003G139500, SORBI_3001G523200 and SORBI_3002G390100 showed high level of expression in all the selected tissues. Drought response of the 11 sorghum genes were analyzed manually by using the dataset GSE80699 and it was observed that 6 genes viz., SORBI_3003G139500, SORBI_3003G024400, SORBI_3001G523200, SORBI_3004G049500, SORBI_3003G302800 and SORBI_3010G186300 were up regulated and 5 genes viz., SORBI_3002G034700, SORBI_3003G339700, SORBI_3005G012000, SORBI_3008G032000 and SORBI_3002G390100 were down regulated during the treatment.
It was identified that above 80% of the genes in respective groups have shown medium or high level expression up on drought stress. It was also noted that all the genes had high or medium level expression at leaf and shoot part. This expression analysis indicates that these orthologous genes posses similar expression with respect to drought stress. The similar expression of orthologous genes are already reported by Kong, et al. [39].
Conclusion
The present study screened 49 genes and scrutinized 31 genes in rice, maize and sorghum for potential drought stress response based on functional analysis. The experimentally proved drought tolerant genes of CIPKs from rice and their orthologous genes in maize and sorghum were grouped by phylogenetic analysis. Maximum number of rice orthologs was found in maize. Comparative analysis of the gene structure showed that Group II (B) CIPK genes dominated intronless feature whereas, Group II (A) CDPK genes and Group I CIPK genes dominated intron rich feature. The intron richness indicates that the genes might have included in multiple stress signal transduction other than drought. The genes in Group II (B) are specifically induced in drought due to their intronless feature [36]. Alternate splicing and exon shuffling could be the reason for functional diversity among CIPK groups [35]. This also points to the adaptation of plants with respect to environmental changes during evolution which in turn altered their phenotypes significantly by transforming the form and function of genes [40]. The most common motifs seen among the groups were parts of protein kinase domain. The similar distribution of motifs in Group I and Group II (A) indicates the functional similarity of the groups. Gene expression analysis showed that above 80% of the genes in respective groups have shown medium or high level expression up on drought stress. The similarity in the expression pattern also shows their functional similarity [39] and conservation of functions between species.
References
- Trenberth KE, Aiguo D, Schrier G, et al. (2014) Global warming and changes in drought. Nature Climate Change 4: 17-22.
- Feng S, Hu Q, Huang W, et al. (2014) Projected climate regime shift under future global warming from multi-model, multi-scenario CMIP5 simulations. Global and Planetary Change 112: 41-52.
- Roche J, Hewezi T, Bouniols A, et al. (2007) Transcriptional profiles of primary metabolism and signal transduction-related genes in response to water stress in field-grown sunflower genotypes using a thematic cDNA microarray. Planta 226: 601-617.
- Roche J, Hewezi T, Bouniols A, et al. (2009) Real-time PCR monitoring of signal transduction related genes involved in water stress tolerance mechanism of sunflower. Plant Physiol Biochem 47: 139-145.
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58: 221-227.
- Sanders D, Brownlee C, Harper JF (1999) Communicating with calcium. Plant Cell 11: 691-706.
- Luan S, Kudla J, Rodriguez M, et al. (2002) Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14: 389-400.
- Sanders D, Pelloux J, Brownlee C, et al. (2002) Calcium at the crossroads of signaling. Plant Cell 14: 401-417.
- Das R, Pandey GK (2010) Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr Genomics 11: 2-13.
- Albrecht V, Ritz O, Linder S, et al. (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca 2 +- regulated kinases. EMBO l5: 1051-1063.
- Kolukisaoglu U, Weinl S, Blazevic D, et al. (2004) Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43-58.
- Kanwar P, Sanyal SK, Tokas I, et al. (2014) Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 56: 81-95.
- Chen X, Gu Z, Xin D, et al. (2011) Identification and characterization of putative CIPK genes in maize. J Genet Genomics 38: 77-87.
- Weinl S, Kudla J (2009) The CBL-CIPK Ca 2+ -decoding signaling network: Function and perspectives. New Phytol 184: 517-528.
- Yang W, Kong Z, Omo-Ikerodah E, et al. (2008) Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). Journal of Genetics and Genomics 35: 531-543.
- Reddy AS, Ali GS, Celesnik H, et al. (2011) Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010-2032.
- Huang C, Ding S, Zhang H, et al. (2011) CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci 18: 57-64.
- He L, Yang X, Wang L, et al. (2013) Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem Biophys Res Commun 435: 209-215.
- Xiang Y, Huang Y, Xiong L, et al. (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144: 1416-1428.
- Thompson JD, Higgins DG, Gibson TJ, et al. (1994) CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Re 22: 4673-4680.
- Kumar S, Stecher G, Li M, et al. (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547-1549.
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275-282.
- Bailey TL, Boden M, Buske FA, et al. (2009) MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: 202-208.
- Jones P, Binns D, Chang HY, et al. (2014) InterProScan 5: Genome-scale protein function classification. Bioinformatics 30: 1236-1240.
- Gebali SE, Mistry J, Bateman A, et al. (2019) The Pfam Protein Families Database in 2019. Nucleic Acids Res 47: 427-432.
- Gasteiger E, Gattiker A, Hoogland C, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784-3788.
- Hruz T, Laule O, Szabo G, et al. (2008) Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics.
- Barrett T, Troup DB, Wilhite SE, et al. (2011) NCBI GEO: Archive for functional genomics data sets-10 years on. Nucleic Acids Res 39: 1005-1010.
- Parkinson H, Sarkans U, Kolesnikov N, et al. (2011) Array express update-an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res 39: 1002-1004.
- Hrabak EM, Chan CW, Gribskov M, et al. (2003) The arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666-680.
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105-132.
- Kipling (2012) Principles of Phylogenetics, Integrative Biology 200A, University of California, Berkeley.
- Lynch M (2002) Intron evolution as a population-genetic process. Proc Natl Acad 99: 6118-6123.
- Roy SW, Gilbert W (2006) The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat Rev Genet 7: 211-221.
- Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet 11: 345-355.
- Zhu K, Chen F, Liu J, et al. (2016) Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci Rep 6: 28225.
- Knighton DR, Zheng JH, Ten Eyck LF, et al. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253: 407-414.
- Yin X, Wang QL, Chen Q, et al. (2017) Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Turnip (Brassica rapa var. rapa). Front. Plant Sci 8: 1191.
- Kong X, Wei LV, Jiang S, et al. (2013) Genome-wide identification and expression analysis of calcium-dependent protein kinase inmaize. BMC Genomics 14: 433.
- Rensing SA (2014) Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol 17: 43-48.
Corresponding Author
Dr. Merlin Lopus, Community Agro Biodiversity Center- MS Swaminathan Research Foundation, Kerala, India
Copyright
© 2020 Lopus M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.