Emerging and Reemerging Human Viral Diseases
Abstract
The pathogens including viruses, parasites and other microbes, are known to emerge and evolve since thousands of the years ago. Viruses are potent infectious agents causing numerous life-threatening diseases in human beings and co-evolved with human evolution. Emerging viruses, especially the RNA viruses are more pathogenic because there is no herd immunity for most humans. The RNA viruses have capabilities to adapt to the rapidly changing global and local environment due to the high error rate of their polymerases that replicate their genomes. They achieve the high evolution rate though through mutation (e.g. Dengue viruses), reassortment (Influenza viruses), and recombination (polioviruses). The best example of emergence and reemergence of viruses is influenza A viruses (e.g. H1N1 and H5N1) which have resulted in many outbreaks, epidemics, and pandemics across the globe. The emergence and reemergence of novel pathogens are associated with the complicated host-pathogen environment and microbe co-evolution with their host. Emergence/reemergence of human viral disease is a never-ending challenge, affecting the social and economic development of a country.
Keywords
Emergence, Reemergence, Human viral disease, Ebola virus, Influenza virus, Vaccine, ZIKA virus, Enterovirus
Introduction
An emerging infectious pathogen may be defined as an etiological agent that has recently appeared within the population or its geographical ranges are rapidly expanding, which are very old phenomena and documented by Persian and Roman philosophers. They wrote about the devastating disease plague in the time of ancient Greeks [1,2]. The most fatal and unforgettable natural disaster in human history occurred in 1918 due to the outbreak of 'Spanish flu' that resulted in the killing of 50-100 million people (3-5% of the world population) across the world [3,4]. In 1956, the influenza pandemic was repeated as 'Asian flu' and caused nearly 2 million deaths. Furthermore, in 1968, the 'Hong Kong flu' resulted in 1 million deaths, and in 2009 recently reemerged 'Swine flu' killed 18,000 people. Taken these together, flu pandemics represents the classical example of emerging RNA viruses. The first case of 'Bird flu' in human was identified in 1997 in Hong Kong and later it spreads to other regions of Asia pacific, Africa, Europe and Latin America. Very recently in 2013, a novel avian influenza a strain H7N9 has emerged in China.
Factors Responsible for Virus Emergence, Reemergence and Adaptation
Viruses, particularly RNA viruses, have high potential to adapt rapidly to a new host and changing environments than other pathogens. Yellow fever virus (YFV) was the first human virus discovered in 1901. Till now, we have accumulated an enormous amount of knowledge about host-virus interactions. However, this understanding is still expanding. One of the main viral adaptation mechanisms is mutation and mostly occurred in RNA viruses. In RNA viruses, RNA-dependent RNA polymerase is an error-prone and does not have any mechanism to correct mutations caused by viral RNA polymerase [5]. The mutation rate in RNA viruses is 10-4 to 10-6 per bp per generation [6]. These mutations may be deleterious or advantageous for virus growth or help viruses in adapting to a new host or changing environment. Studies have shown that mutations are important for the virulence in case of foot and mouth disease and polio viruses [7].
The other adaptation mechanisms include reassortment, and recombination. Influenza virus evolved and changes their genes by reassortment. When the same host is infected by two different strains of influenza viruses (e.g. avian and human viruses), a new progeny virus with mixed genetic materials could appear. This new strain of influenza virus arose due to reassortment process could be more effective to evade to a new host that lacks immunity to this new pathogen. The emerging Influenza strains which appeared in influenza pandemics in 1957, 1968 and 2009 were raised through this process of gene shuffling by reassortment [8].
Recombination could also produce new strains of viruses. In this case, if two similar viruses infect same host cells and exchange their genetic material in different regions of their genome, a new strain of virus could be generated. This mechanism of recombination mostly occurs in enteroviruses which infect the gastrointestinal tract for long period of time [9]. Another good example of this mechanism is polio live vaccine which contains 3 strains of polio viruses. These strains not only undergo mutations but also recombine among themselves to reverse neuropathogenicity [10,11].
Besides genetic mechanisms, other factors that contribute to the emergence and reemergence of infectious disease are: 1) Ecological changes and agricultural development; 2) Changes in human demographics and behavior; 3) Migration of human across different geographical region and commerce; 4) Breakdown of public health measures and deficiencies in public health infrastructure; 5) Climate change also plays important role in the emergence of an infectious disease; 6) Resistance to microbial antibiotics; and 7) Zoonosis (from animal to human). Emerging and reemerging human viruses have summarized in Table 1. This review is focused on emerging RNA viruses that infect humans and may cause deadly disease. Taken together, we are focused on the emergence and reemergence of ZIKA, Dengue Virus, West Nile Virus (WNV), EBOLA, Influenza, SARS-CoV, MERS-CoV, Nipha Virus, and enteroviruses.
Zika virus
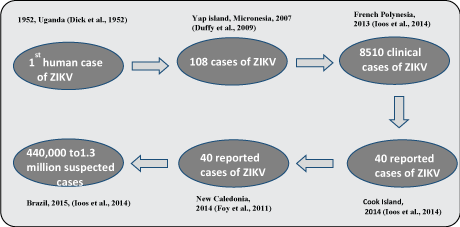
Zika virus (ZIKV) is a flavivirus and primarily transmitted by the bite of an infected mosquito. ZIKV is related to other flaviviruses like dengue virus, West Nile virus and yellow fever virus. The recent emergence and spread of ZIKV to 48 countries across worldwide have been considered as an emerging global public health problem. At present, there is no effective vaccine to prevent ZIKV infection. ZIKV is transmitted by mosquito bite was confirmed when it was first isolated from Aedes aegypti mosquitoes in 1966 in Malaysia [12]. The ZIKV resides in the salivary gland of infected mosquitoes and inoculated into the human body by mosquito's bite during their blood feeding. ZIKV is also found to be transmitted sexually from one person to another as the ZIKV RNA has been observed in sperm and urine after 62 days of infection [13-15]. ZIKV is an enveloped, spherical virus containing single-stranded positive RNA genome with 10,794 nucleotides. This RNA is translated into a poly protein which is co & post-translationally cleaved into 11 mature viral proteins. ZIKV infection in Asia was observed in Indonesia where seven persons were suffering from fever, stomach pain, dizziness and anorexia [16,17]. The first significant outbreak of ZIKV infection occurred in 2007 in Yap Island of Micronesia [16]. A total of 59 persons were infected by ZIKV as confirmed by reverse-transcription polymerase chain reaction (RT-PCR) and serological analysis. Figure 1 summarizes the outbreaks of ZIKV in human population in different countries. It was found that up to 73% of Yap Island residents were infected by ZIKV, and Aedes hensilli was considered as a principal vector for ZIKV transmission. The second major ZIKV outbreak emerged in French Polynesia in 2013 where 28,000 of people were supposed to be infected with ZIKV based their signs and symptoms of low-grade fever, conjunctivitis, arthralgia and maculapapular rash [18]. This is the first time, Guillain-barre's syndrome was linked with the ZIKV infection.
In 2015, Brazil informed the WHO about ZIKV infection. ZIKV infection in Brazil leaded to 440,000 to 1.5 million cases and represents an emergency threat to human life especially to pregnant women and newborns [17,19]. The first case of ZIKV infection in South America was confirmed in May 2015. It is assumed that ZIKV was introduced into the country through the World Sport Championship, the canoe race held in Rio de Janeiro in August 2014. Four countries, French Polynesia, New Caledonia, the Cook Islands, and Easter Island were participating in this championship. ZIKV strain in Brazil was closely related to the strain in French Polynesia outbreak in 2013-2014. ZIKV congenital infection causes microcephaly in newborns and its infection is related to Guillain-barre syndrome in adults. ZIKV infects and replicates in neural stem cells of developing brain and causes their apoptosis that leads to the reduction of cortical layer thickness in a developing brain [20]. In America, the increasing number of cases of congenital microcephaly and Guillain-Barre syndrome cases are related to ZIKV infection [18,21,22]. On 1st Feb 2016, WHO declared ZIKV infection as an international health emergency. ZIKV is able to cross the placenta, detectable in amniotic fluid and causes intrauterine growth restriction of a fetus in pregnant women [20,23,24]. Three lineages of ZIKV have been identified by phylogenetic analysis. Two are African lineages, e.g. MR766 and Nigerian cluster and one is Asian lineage (Table 2) [25].
Most of the ZIKV infection is asymptomatic and incubation period is not known precisely but usually the cases occur 3-13 days after a mosquito's bite [16]. Clinical symptoms of ZIKV infection include mild fever, conjunctivitis, skin rashes, joint and muscle pain, back pain and headache. Other clinical manifestations are anorexia, dizziness, diarrhea, constipation, retro-orbital pain, edema, and abdominal pain. ZIKA vaccine including DNA vaccine, neutralizing antibody vaccine, subunit vaccine and others are in progress [26,27].
The ZIKV infection in infected people can be detected by RT-PCR and enzyme-linked immunosorbent assay (ELISA) using IgM antibodies or plaque reduction neutralization test (PRNT). IgM cross reacts with other flavivirus. Center for Disease Control and Prevention recommends to use both RT-PCR and serological test to confirm ZIKV infection.
West Nile Virus (WNV)
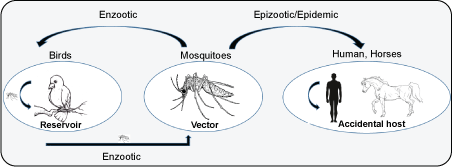
West Nile Virus (WNV) is an emerging virus belongs to the family Flaviviridae, genus Flavivirus. WNV is a single-stranded, positive sense RNA virus with a genome of approximately 12 kb. WNV was first isolated in the West Nile district of Uganda in 1937 [28]. WNV is transmitted to humans by mosquito bite and to some animals like birds. Figure 2 summarizes the WNV transmission cycle in human and birds. Birds serve as the reservoir of the virus. The infection of WNV in humans is usually asymptomatic (80%) and caused West Nile fever and encephalitis.
WNV is a neurotropic flavivirus infects the central nervous system and causes severe neurological diseases [29]. The symptoms of West Nile neuroinvasive disease (WNND) include the encephalitis, meningitis, and acute flaccid paralysis. WNV enters the central nervous system and infects neuron, glial cells, basal ganglia, spinal cord and brain stem [30]. The infected neurons and astrocytes produce the chemokines, CXCL10 and CCL5. These chemokines play important role in WNV specific T-cells into CNS [31]. During the period from 1937-1999, the WNV infection in humans, horses, and birds occurred occasionally [32,33]. However, a small outbreak of WNV infection in humans and horses occurred in Camargue region in France in 1960s [34]. WNV was first introduced in the Western Hemisphere in 1999. Infection in humans caused 62 cases, 59 cases with neuroinvasive and 7 deaths in New York City [35]. WNV epidemic is usually characterized by the introduction of few human cases in first season with explosive amplification in second season and consequence maintained of level. WNV is a neurotropic flavivirus infects the central nervous system and causes severe neurological diseases [29].
Sequence analysis shows that WNV in New York City was imported from Israel [29]. WNV was first detected in Florida in 2001 indicating that virus had spread to other parts of country. Later in 2001, the virus was detected in 21 states in America including Iowa and Louisiana with 64 neurotropic disease cases and 9 deaths [36]. The virus continues to spread to other parts of the country, reach to California and caused largest epidemics of meningoencephalitis in history of country [37]. According to the national surveillance in America, the total number of WNV case and deaths from 1999-2013 were 37,090 and 1489 respectively [38]. Initial cases of WNV infection in human around the world has summarized in Table 3.
Most of the people (70-80%) infected with WNV do not show symptoms of infection. Less than 1% of infected people show some neurological symptoms like encephalitis or meningitis. The symptoms of WNW infection in humans includes fever, body ache, headache, vomiting, diarrhea, skin rash, coma and fatigue. Diagnostic test for WNV include the blood test to check raising level of antibodies against WNV, brain test that measure the brain activity by electroencephalography (EEG) to check brain inflammation by WNV and lumbar puncture [39]. Currently there is no vaccine or antiviral treatment of WNV infection [40].
Dengue virus
Dengue is a common viral disease causing 50 million cases every year in more than 100 countries in tropical and subtropical regions. Dengue virus (DENV) belongs to genus flavivirus and family Flaviviridae. Dengue is considered as second largest mosquitoes borne disease after malaria. The majority of dengue virus infections are asymptomatic however minority of cases leads to dengue hemorrhagic fever (DHF), dengue shock syndrome (DSS) characterized by circulatory failure. Dengue virus infection in human has been also associated with neurological symptoms [41]. Dengue virus is a positive-sense, single-stranded RNA virus with a genome of 11 kb. Dengue virus exists as four antigenically distinct serotypes named DENV 1-4. Dengue virus is transmitted to human by the bite of mosquitoes, Aedes aegypti during their blood meal. The symptoms of dengue virus infection are majorly asymptomatic therefore it is very difficult to predict first dengue infection in humans. According to a Chinese medical encyclopedia, first dengue infection might be dated to 992 A.D. [42]. It is very difficult to predict place of origin of dengue virus but some studies show that this virus might have originated in Africa as many mosquitoes-borne diseases occurred there and often infect primates [43]. Another study predicts the Asian origin of dengue virus [44]. By late 19th and early 20th century, the virus widespread to tropical and subtropical countries [45]. First reported outbreak of dengue virus occurred in 1953/1954 in Manila and the second outbreak occurred in 1958 in Bangkok [46]. Sudden outbreaks of dengue in China from 1978-2008 have resulted in more than 600,000 cases approximately and about 475 deaths overall [47,48]. As far as America is concerned, DENV is expanding its geographical range and reintroduce in North America through Texas and Florida in America. The DENV epidemic is limited by a control campaign initiated in 1947 by Pan America Health Organization (PAHO). This organization is focus on removing the Aedes aegypti from Central and South America. The PAHO program was suspended in 1970s and Aedes aegypti population increased again that leads to the raise in DENV cases [49]. DENV outbreaks in 2009 in Florida occurred first in more than 70 years and 16 cases of DENV were reported. In 2010, DENV causes infection in 66 people in Florida and after that few cases of DENV infection reported [50,51]. According to a CDC report in 2013, 4 cases of dengue virus infection in Hawaii in 2011 and 2 cases in Florida in 2012 were reported. Moreover, in 2013, 20 cases of dengue in Florida and 3 cases in Texas were reported [52]. Very recently, according to a newspaper, a total of 80 cases of dengue have been reported in USA and its territories in 2017 [53]. It indicates that the DENV is increasing its geographical ranges and may causes pandemics in near future.
Ebola virus
Ebola virus (EBOV) is the deadliest emerging virus causing infection in human and nonhuman primates (NHP). It is a filovirus and causes hemorrhagic fever in human. It is an enveloped, single-stranded, non-segmented, negative RNA virus. EBOV virus genome is approximately 19 kb and encodes for 7 genes resulting 9 proteins. EBOVs together with Marburg viruses constitute the Filoviridae family [54]. There is no FDA-approved drug or vaccine to treat Ebola virus infection in human. The only way to control the disease is to keep the patients in isolation, protect the health care workers and close monitoring of the people who come in contact with the patients. In Africa, bats serve as a reservoir for EBOV transmission [55]. Human get infected with EBOV when they come in contact with infected tissues, blood, bats, patients or accidental host (Apes). EBOV was first identified in 1967 in Marburg, Germany when a few animal workers and staffs got infected. These peoples were processing samples from monkey imported from Uganda to prepare kidney cells to study Poliomyelitis vaccine [56,57]. The first significant outbreak of Ebola was recognized in 1976 when it caused hundreds of deaths in Norther Zaire (318 cases) and southern Sudan (284 cases) [38]. Among these two subtypes, Zaire subtype of Ebola virus was more fatal and resulted in 90% of case fatality while Sudan type causes 50% of case fatality. In 1979, EBOV erupted again and caused infection in 34 people in Africa and 12 deaths [58]. Three more species were identified in subsequent EBOV infections. In 1989, Reston EBOV was discovered which causing infection in Cynomolgus Macaques imported from the Philippines [58]. There was no proof that it could also cause infection in humans [59]. In decades of 1980s and 1990s, a very few cases of symptomatic EBOV infection in human were observed until 1994. In 1994, 31 people were killed due to EBOV infection in Minkoka district in Gabon [60]. After that, the EBOV outbreaks occurred every 1-4 years in Sudan, Congo, Sudan and Gabon.
Another major outbreak of EBOV occurred in 2007 in Uganda where 149 cases with 25% fatality were observed. Furthermore, another outbreak of EBOV occurred in December 2013 in Guinea and quickly spread to other adjacent countries Sierra Leone and Liberia. It was considered as the largest outbreak in history. Zaire strain of EBOV responsible for this outbreak was named as Makona after the river which is present at the borders of these three countries. The EBOLA continue to emerge, and epidemic accelerated to other countries in Africa. As a result, a total number of 2347 cases were identified in central Africa till 2013 [38]. In August 2014, an unrelated outbreak of ZEBOV occurred in the Democratic Republic of the Congo and caused 66 cases with 74% lethality rate. By the March 2016, a total of 28,646 cases of EBOV disease and 11,323 deaths have been reported across 10 countries (Liberia, Guinea, Sierra Leone, Italy, United Kingdom Mali, Senegal, Spain, Nigeria and United States) [61].
The incubation period of EBOV disease ranges from 2-21 days. The symptoms of the disease are fever, chills, nausea, vomiting, anorexia, abdominal pain and diarrhea. Death is caused by multiple organ failure and shock. EBOV infection in human takes place via entry through the mucous membrane, injuries, and abrasion in the skin or parental transmission. EBOV can invade in almost all the cells of body by a different mechanism for each cell type. Usually, EBOV enters into cells via lipid rafts, endocytosis and macripinocytosis [62,63].
During early days of infection, it is difficult to diagnose EBOV disease as the symptoms are more common to other diseases like malaria and typhoid fever. EBOV is detected in blood only after onset of infection. Table 4 summaries the few diagnosis methods of EBOV disease.
Influenza virus
Influenza virus is another emerging virus which belongs to the Orthomyxoviridae family and infects many species. Influenza virus contains negative-stranded RNA genome with 8 segments and is an enveloped virus. Segmented genome allows the viruses reassortment which means that if two viruses infect the same cells then progeny viruses may have gene segments from both the parents with novel antigenic and genetic properties. The reassortment between viruses allows virus adaptation to a new host and raises the potential to cause pandemics [64].
Influenza virus encodes two spike-shaped surface proteins named hemagglutinin (HA) and neuraminidase (NA). The HA required for virus attachment to target cells [65] and NA allows the release of progeny viruses from infected cells. The influenza A viruses are classified based on their subtype of hemagglutinin (H1-H17) and neuraminidase (N1-N10) [66,67]. Seasonal influenza viruses kill 250,000-500,000, mostly old people (90%) each year around the world [61]. The main reservoir for influenza viruses is wild waterfowl. Thus, they serve as the main source for emergence and reemergence of influenza viruses. Avian influenza can be highly pathogenic and continue to emerge and reemerge around the globe to cause many epidemics and pandemics. Avian influenza virus was existed as 'fowl plague' and known more than 130 years. Chicken, ducks, and pigs also serve as the reservoirs of influenza virus and act as a mixing vessel for the emergence of novel strains of influenza virus that not existed previously. The first instance of 'Fowl plague' was first recognized in 1878 in Northern Italy [68]. The most fatal and unforgettable natural disaster in human history occurred in 1918 due to the outbreak of 'Spanish Flu' that resulted in the infection of 500 million people across the world and killing of approximately 100 million people [4].
H1N1 influenza
The H1N1 influenza virus has caused many significant mortalities and morbidities in the human population. An influenza pandemic in 1918 was caused by H1N1 virus. Segmental reassortment plays important roles in the evolution of H1N1 virus. The 1918-1919 Influenza pandemic during First World War resulted in half million deaths in United State and was caused by H1N1 [3]. The pigs were considered as the mixing vessels for this influenza pandemic [69]. The influenza pandemic was not reported before, therefore soldiers and people were not aware and could not take preventive actions. The U.S. imposed the quarantine to prevent the spread of virus as people were dying in large number and dead bodies were mounted up.
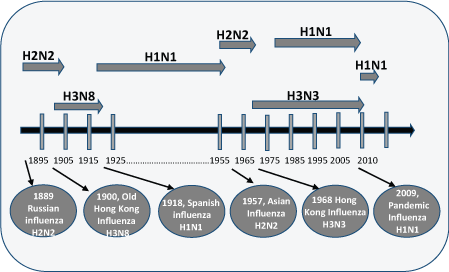
After 1918 influenza pandemic, severe outbreaks of H1N1 occurred in 1928-1929, 1932-1933, 1936-1937 and 1943-1944 in the United States [70] and the United Kingdom [71]. Intrasubtypic reassortment of H1N1 occurred in 1947 and this new subtype was not neutralized by the existing vaccine and caused more severe disease. The virus continued to spread across the globe and named as 'A-prime' based on antigenic divergence [72]. In this subtype, the HA segment was mutated whereas NA segment remained conserved [73]. The H1N1 disappeared from human in 1957 and a new strain, H2N2 appeared. This new strain contains three segments from avian source and five segments from H1N1 1918 strain lineage [74]. After the emergence of this new subtype, H1N1 did not appear until 1977 [75]. The influenza A (H1N1) reemerged in 1977 in North Eastern China, Soviet Union, and Hong Kong, and mostly affected young people [74]. This new strain was closely related to the strain from 1950 indicated that this strain was existed and did not disappear. Figure 3 summarizes the emergence of influenza virus in human population at different time points.
Reassortment between different influenza strains isolated from different hosts species leads to the generation of new strains having a potential for global pandemic. The best example for this is the reassortment between avian and human influenza produced H2N2 and H3N2 that caused pandemic in 1957 and 1968 respectively [74,76]. Recently, it has been shown that reassortment between influenza a viruses among H3N2 subtypes leads to the production of novel influenza virus [8]. Influenza pandemics occurred in humans have listed in Table 5.
Swine influenza
Swine influenza also called as hog flu, pig influenza or pig flu. Swine influenza virus infects pigs and usually does not cause infection in humans. However, they do pose significant threat to people working with the pigs. H1N1 swine flu is circulating in pigs in North America and other regions since past 80 years [77]. In 1997, a highly pathogenic avian virus H5N1 caused infection in humans in Hong Kong with 18 confirmed cases and 6 deaths. This was an unusual case where avian virus caused infection in humans and had never reported before. In 1998, a novel strain of H3N2 emerged in North America and caused an outbreak in North American swine [78]. Furthermore, in 2009, a novel swine-origin influenza A (H1N1) was identified and caused infection in human [79]. This new strain of swine origin influenza A virus (S-OIV) arose from triple reassortment and contain genes from human H3N2, classical swine H1N1, and North American avian influenza viruses. The human to human transmission of S-OIV and other influenza viruses is thought to occur through the droplets released during coughs. In United States, a total of 642 confirmed cases were identified in human from 2005-2009 [79]. Moreover, the mixing and co-circulation of H3N2 further generated H1N1 and H1N2 which caused sporadic infection in humans [80,81]. H1N1 entered in Italy when a shipment of pigs from United States introduced into Italy in 1976. This strain rapidly spreads among other swine. It was shown that a new avian-origin influenza A strain introduced in European pigs from Ducks [82]. A new strain of swine-origin influenza A (H1N1) emerged in Mexico and the United States in 2009 [83]. This virus spread rapidly to 30 countries via human to human transmission mode and WHO declared pandemic alert of level 5 of 6. According to WHO, the pandemic alert of level 5 characterized by human to human transmission of virus in at least two countries in a single WHO region while level 6 characterized by community level outbreaks in different WHO regions in addition to phase 5 level criteria. This new strain of influenza virus supposed to have potential to cause influenza pandemic in twenty-first century.
Common symptoms of influenza include fever, nasal congestion, headache, lack of energy, sore throat, runny nose, and muscle aches. The symptoms of influenza vary from person to person depending on the immune system, age, and medical conditions. The diagnostic tests for influenza include the serology, virus culture, RT-PCR, rapid molecular assays, rapid antigen testing and immunofluorescence assay (IFA) [84].
Influenza vaccines also known as influenza shots developed twice a year as influenza virus changes very rapidly. WHO and CDC recommend the influenza vaccination in nearly all the people above the age of 6 years. The vaccination effectiveness changes year to year and provides modest to high protection against influenza.
Hong Kong summer influenza, 2017
A recent unusual outbreak of summer influenza in Hong Kong has been reported in May 2017. In Hong Kong, May 2017 to August 2017, summer influenza outbreak has caused a total of 14,713 cases and resulted in 306 deaths (Avian Flu Diary, Aug 2017) Table 6.
This flu is considered as unusual in many aspects: a) It started one month earlier than two previous years; b) It caused over 300 deaths, which is much higher than last summer; c) It has sustained more than 90 days as compared to few days in last year. This outbreak may be due to the flu vaccine administered last year has begun to expire and may not be effective in providing protective immunity to the vaccinated people. It may also be due to weak flu peak last year and thus the number of people developed natural immunity would be less. This outbreak may also be due to the mutation of a dominant antigenic epitope within the H3N2 flu strain, making the flu vaccine used in the past two years ineffective [85].
Severe Acute Respiratory Syndrome Corona virus (SARS-CoV)
Severe acute respiratory syndrome (SARS) is another emerging corona virus and considered as a threat to human health. SARS-CoV transmits from one person to other through the droplets produced during a cough or sneezing, direct contact with infected droplets. SARS virus replicates in lungs and gastrointestinal tract [86]. There is no vaccine or antiviral drugs to treat SARS. The SARS is caused by a corona virus. The first case of SARS was recorded in city of Foshan, Guangdong Province, China in November 2002. The Chinese Ministry of Health reported to WHO about an 'acute respiratory syndrome' outbreak causing 300 infected cases and 5 deaths. The symptoms were similar to atypical pneumonia. A 64-year-old doctor got infected with the SARS while he was treating SARS patients. This doctor was traveling to Hong-Kong and stayed in Metro pole Hotel, where 16 other people got infected with SARS. These people carried the virus to Singapore, Toronto and Hanoi. The doctor got very sick on following day and died on March 4 in a hospital. On March 15, WHO issued a global alert about the new emerging respiratory disease, SARS. After that SARS spread to 36 countries in Asia, Europe, and North America within next 6 months. Center of origin for SARS is believed to be Guangdong province in Southern China. A total number of 8096 cases were reported and 774 patients died of infection [87-89]. The symptoms of infected people were similar to influenza and include headache, fever, myalgia, respiratory failure, and death. In 2003-2004, SARS reemerged and infected 4 people. These four people were recovered [90,91]. The animals, Himalayan palm civets, and raccoon dogs were considered to harbor virus very similar to SARS [92].
Middle East Respiratory Syndrome Corona virus (MERS-CoV)
After 2004, no more case of SARS reported, however, another similar disease called Middle East respiratory syndrome (MERS) continue to appear. The MERS caused by a corona virus. The first case of MERS was reported in Saudi Arabi in June 2012 [93]. MERS cases were also detected in Qatar, Oman and the United Arab Emirates [94]. MERS rapidly spread to other countries in Asia, Europe, and Africa by travelers resulting in total 1626 people infection with 36% fatality till Jan 2015 [95].
Nipah Virus (NIV)
Nipah virus is a recently discover non segmented RNA virus belongs to paramyxovirus family. It is a highly pathogenic virus and laboratory work requires bio safety level-4 containment. Nipah virus enters into central nervous system (CNS) and causes encephalitis in infected individuals with a mortality rate of 40-70%. However, the exact route of entry of Nipah virus into CNS is unknown but study have shown that it could enters through the olfactory epithelium in nasal turbinates [96]. The genome of NiV consists of six genes (N-P-M-F-G-L) flanked by 5'trailer region and 3'leader region [96]. The fruit bats of Pteropus genus considered as the main reservoir of NiV. The NiV can infect many mammalian species like cats, horses, dogs and pigs. NiV has been considered as emerging virus threaten to human health in Southeast Asia. The NiV has no-segmented RNA genome and considered as highly mutagenic virus. NiV caused the deadly disease in human and have wide reservoir hosts [97]. This virus was first recognized in 1998 when it caused viral encephalitis outbreak in Malaysia. This outbreak infected 300 people with 35% mortality [98]. This outbreak was caused by NiV transmission from pigs to human [99] and culling of 1 million pigs leads to the control of outbreak. Initially, it was considered that the outbreak was caused by the Japanese encephalitis (JE) which is a mosquito born RNA virus, but JE vaccination could not treat the disease. After initial outbreak, NiV has reemerged in Bangladesh in 2001, 2003, 2004, 2005, 2007 and 2008 that caused fetal human encephalitis [100,101]. NiV has also reemerged in India in 2001 and 2007 that also leads to fatal human encephalitis. NiV infection can be diagnosed by virus isolation and RT-PCR from nasal and throat swab, urine, cerebrospinal fluid and blood. Antibody detection (IgG and IgM) can also be used to detect NiV infection. Nosocomial infection of NiV can be prevented by standard control practice and proper nursing techniques. The ribavirin drug is effective in vitro but clinical application of ribavirin remained uncertain.
Human Enterovirus
Human enteroviruses have caused many epidemics in recent history and considered as important emerging viruses infecting humans mainly infants and children. So far, only humans seem to be host of the enteroviruses. Enteroviruses are positive-sense, single-stranded RNA viruses and belong to genus enterovirus and family picornaviridae. The RNA genome is enclosed in an icosahedral capsid. Human enteroviruses are classified mainly into three classes, EV-A, EV-B, EV-C, and others as D-J [102,103]. The human enteroviruses Enterovirus A species consist of 25 serotypes. Enteroviruses responsible for hand-foot-and-mouth disease (HFMD), myocarditis, herpangina, pleurodynia and meningitis. The EV-A71 and Coxsackievirus A16 are two of the enterovirus A viruses that caused major outbreaks recently.
Enterovirus-A71 (EV-A71)
EV-A71 is one of the hundreds of enteroviruses, has infected millions of people, and associated with life threatening complications in newborns. EV-A71 causes HFMD, herpangina, cerebellitis, meningoencephalitis, and has circulated in Asia-pacific since 1997. EV-A71 is divided into six subgroups, only B and C subgroup are associated with outbreaks [104]. Usually, EV-A71 is more pathogenic as compared to other enteroviruses, and causes neurological complication. EV-A71 was first discovered in California, USA, where it caused several outbreaks and found to be associated with neurological symptoms in 1969-1972 [105]. EV-A71 outbreaks have occurred in many countries and regions, including China, Australia, Cambodia, Vietnam and Taiwan. In China, the first large outbreaks of EV-A71 occurred in 2007 in Shandong Province where 1149 cases and 3 deaths were reported [106]. Furthermore, in 2008, EV-A71 caused a major outbreak of HMFD in Fuyang city, China. A total of 6049 cases were reported with several fatalities [107]. Another epidemic of EV-A71 occurred in Guangdong Province in China where it caused 48,876 cases (131 cases were severe and 21 cases were fatal) [108]. Therefore, EVA-71 has become an important problem for China. The main reasons for EV emergence are the mutations and recombination. The infidelity of polymerase leads to one mutation per genome pre-replication [109]. In Vietnam, a total of 63,780 cases of HFMD were reported in 2012 and of which, approximately 58% were caused by EV-A71 [110].
In Europe, EV-A71 outbreaks have caused many epidemics in history. The first major outbreak of EV-A71 in Europe occurred in 1975 in Bulgari where it infected more than 705 people, of which 149 developed paralyses and 44 died. Recently in France, EV-A71 and EV-D68 have caused a total of 59 cases for severe pediatric conditions in 2016 [105]. Fifty-two of children were affected by severe neurological symptoms. Another outbreak of EA-A71 in Catalonia in 2016 caused hundreds of the cases affecting children up to the age of 10 and associated with neurological symptoms [111]. These outbreaks of enteroviruses give us a warning about more epidemic in the near future that may be fatal to humans.
EV-A71 spread through the fecal-oral route, by respiratory droplets, oral secretion, and fomites. The most sensitive and rapid diagnostic test for EV-A71 is RT-PCR performed on clinical samples including blood, cerebrospinal fluid, stool or urine [112].
Enterovirus-D68 (EV-D68)
EV-D68 is an emerging virus causing many outbreaks in recent history. EV-D68 was first reported in 1962 in California where four children were diagnosed with pneumonia and bronchitis. In recent history, the increasing numbers of respiratory diseases caused by EV-D68 have been reported in many countries including the United States, the Netherlands, Philippines, and Japan [113]. During 1970-2005, only 26 cases of EV-D68 were identified and the majority of the cases were in children [114]. Small and geographically limited outbreaks of EV-D68 occurred globally during 2008-2010 [113]. During these outbreaks, 11 patients from Japan, 21 patients from Philippines, 24 patients from the Netherland and 39 patients from United States were reported [113]. The subsequent outbreaks of respiratory disease caused by EV-D68 were reported in different countries including Great Britain, Italy, Thailand, China, Italy, New Zealand, and several African countries [115]. The largest outbreak of EV-D68 occurred in the United States and Canada during 2014 [116] and found to be associated with severe clinical manifestation. This outbreak caused significant morbidity and mortality. In kanas City, USA, nineteen patients were found to be infected with EV-D68 in 2014. These patients were ranged from six weeks to 16 years [116,117]. Similar outbreaks occurred in Chicago. According to a CDC reports, a total of 1153 case of EV-D68 infections were confirmed in 49 states including Columbia district [38]. EV-D68 infections were not confined to USA only, the Public health agency in Canada reported of 268 cases of EV-D68 in several cities during 2014 [118], followed by other countries in Europe including Germany, Netherlands, Denmark, Sweden, Spain, France, Italy, and also from Chile and Brazil [119]. Recently in 2016 in France, EV- EV-D68 and EV-A71 have caused a total of 59 cases for severe pediatric conditions associated with different symptoms [115]. The increasing number of EV-D68 infections may be due to the more adaptation of virus, increasing geographical ranges and enhanced virulence. The molecular mechanism behind recent emergences of EV-D68 is considered to be the antigenic variations that leads to generation multiple sublineages, novel variants and loss of protection from preexisting antibodies [119]. The diagnosis of EV-D68 is done by PCR method performed on clinical samples. Recently, a real time PCR (RT-PCR) based method have been implicated in the diagnosis of EV-D68 and this method is very sensitive and specific [120,121].
Prevention and treatment of enteroviruses
There is no FDA approved drug for the treatment of enterovirus infection. Prevention and supportive treatment are the best way to treat the enterovirus infection. The vaccine is available for poliovirus. In 2015, the Food and Drug Administration (FDA) of China have approved the first vaccine for the treatment of EV-A71. This vaccine was developed by the Institute of Medical Biology at Chinese Academy of Medical Sciences. The second vaccine for EV-A71 was approved in 2016 by Chinese FDA and this vaccine was made by Sinovac Biotech [122].
Conclusion
The emergence and reemergence of viral disease are considered as a continuous threat to human life in the era of globalization [123]. The emerging and reemerging viruses include the RNA viruses. RNA polymerase of RNA viruses is error-prone [5]. The mutations in the genetic material of RNA viruses accumulated over the time and produce a new strain of virus with novel immunogenic and antigenic properties. This new strain of virus has a potential to cause a pandemic. In addition to the genetic variation, environmental factors such as changing environment and tropical deforestation also play important role in the emergence of a viral disease [124]. Furthermore, a number of several other factors are responsible for the emergence of a new disease and reemergence of the old viral diseases [124]. Viruses, especially the RNA have the intrinsic property to change their genetic material according to the changing environment and condition [5]. Emerging RNA viruses cannot be conserved in their ecological niche and have potential to cause infection in alternate host species. Rapidly increasing population growth, poverty, international travel by the workers, immigrants, refugee and tourist favor the spread of viral disease across the world.
Recent advances in the whole genome and transcriptome sequencing and structure-based vaccine and drug designed have increased our ability to understand the mechanism of disease emergence. History of an infectious disease can reflect the history of the microbe causing disease. Genome sequencing of emerging virus at different time points of its emergence may tell us the evolution of particular virus at nucleotide and protein level. Viruses have the intrinsic potential to change their nucleotides by mutation, recombination, genome segment reassortment and a combination of these molecular process to produce a new progeny which is phenol typically diverse than parental virus [5]. Mutations in the structural and nonstructural proteins of viruses increase their host range and cause non-human viruses to infect humans. The majority of emerging viral disease is zoonotic in nature and several ecological, sociological and changing environments lead to the emergence of new viruses. The many emerging pandemics caused by Dengue virus and WNV have been controlled by the implementation of various preventive measures, use of mosquitoes' net, decant of stored rain water so that mosquitoes cannot breed, and application of mosquitos' disinfectant. We still do not know the factors that determine the interspecies transmission, reassortment and emergence of new viruses. The factors that responsible for the pandemics in the past may be critical for the emergence of new pandemic in future. The study of virus evolution in a real time may provide us the imperative information regarding the virus pathogenesis, emergence and reemergence.
Prevention of emerging viral diseases: The individuals from various expertise like biologist, chemist, doctors, environmental scientists, and ecologist must work together to understand the mechanism and factors for an emergence of a viral disease or other infectious diseases. The global surveillance of an emerging disease can give early warning about the emerging pathogens. Development of new effective drugs, vaccines including DNA vaccines, subunit vaccine, and other measure control should be explored to control the emerging viral and other infectious diseases. WHO have created a global early warning system (GLEWS) in collaboration with World organization for Animal Health and Food and Agriculture Organization. GLEWS aim to provide the early warning and risk assessment of zoonosis and emerging infectious diseases. GLEWS also provide information about the geographical spread of novel viruses and reemerging viruses. Understanding the factors responsible for the disease emergence and reemergence can help us to develop the effective strategies for the prevention of a disease progression.
Acknowledgements
This work is partially supported by grants from Guangdong Innovative and Entrepreneurial Research Team Program (No.2014ZT05S136).
References
- Krause RM (1992) The origin of plagues: Old and new. Science 257: 1073-1078.
- Morens DM, Folkers GK, Fauci AS (2008) Emerging infections: A perpetual challenge. Lancet Infect Dis 8: 710-719.
- Mills CE, Robins JM, Lipsitch M (2004) Transmissibility of 1918 pandemic influenza. Nature 432: 904-906.
- Taubenberger JK, Morens DM (2006) 1918 influenza: The mother of all pandemics. Emerg Infect Dis 12: 15-22.
- Menendez-Arias L (2002) Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog Nucleic Acid Res Mol Biol 71: 91-147.
- Lauring AS, Frydman J, Andino R (2013) The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11: 327-336.
- Haydon DT, Bastos AD, Knowles NJ, et al. (2001) Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics 157: 7-15.
- Holmes EC, Ghedin E, Miller N, et al. (2005) Whole-genome analysis of human influenza a virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 3: e300.
- Lukashev AN, Shumilina EY, Belalov IS, et al. (2014) Recombination strategies and evolutionary dynamics of the Human Enterovirus a global gene pool. J Gen Virol 95: 868-873.
- Kirkegaard K, Baltimore D (1986) The mechanism of RNA recombination in poliovirus. Cell 47: 433-443.
- Jarvis TC, Kirkegaard K (1992) Poliovirus RNA recombination: Mechanistic studies in the absence of selection. EMBO J 11: 3135-3145.
- Marchette NJ, Garcia R, Rudnick A (1969) Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg 18: 411-415.
- Nicastri E, Castilletti C, Liuzzi G, et al. (2016) Persistent detection of Zika virus RNA in semen for six months after symptom onset in a traveller returning from Haiti to Italy, February 2016. Euro Surveill 21.
- Harrower J, Kiedrzynski T, Baker S, et al. (2016) Sexual Transmission of Zika Virus and Persistence in Semen, New Zealand, 2016. Emerg Infect Dis 22: 1855-1857.
- Mansuy JM, Suberbielle E, Chapuy-Regaud S, et al. (2016) Zika virus in semen and spermatozoa. Lancet Infect Dis 16: 1106-1107.
- Duffy MR, Chen TH, Hancock WT, et al. (2009) Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360: 2536-2543.
- Brasil P, Calvet GA, Siqueira AM, et al. (2016) Zika virus outbreak in rio de janeiro, Brazil: Clinical characterization, epidemiological and virological aspects. PLoS Negl Trop Dis 10: e0004636.
- Cao-Lormeau VM, Blake A, Mons S, et al. (2016) Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet 387: 1531-1539.
- Cerbino-Neto J, Mesquita EC, Souza TM, et al. (2016) Clinical manifestations of Zika virus infection, Rio de Janeiro, Brazil, 2015. Emerg Infect Dis 22: 1318-1320.
- Cugola FR, Fernandes IR, Russo FB, et al. (2016) The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534: 267-271.
- Mlakar J, Korva M, Tul N, et al. (2016) Zika virus associated with microcephaly. N Engl J Med 374: 951-958.
- Martines RB, Bhatnagar J, Keating MK, et al. (2016) Notes from the field: Evidence of Zika virus Infection in brain and placental tissues from two congenitally infected newborns and two fetal losses--Brazil, 2015. MMWR Morb Mortal Wkly Rep 65: 159-160.
- Li C, Xu D, Ye Q, et al. (2016) Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19: 120-126.
- Miner JJ, Cao B, Govero J, et al. (2016) Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165: 1081-1091.
- Gong Z, Gao Y, Han GZ (2016) Zika virus: Two or three lineages? Trends Microbiology 24: 521-522.
- Lagunas-Rangel FA, Viveros-Sandoval ME, Reyes-Sandoval A (2017) Current trends in Zika vaccine development. J Virus Erad 3: 124-127.
- Jaijyan D K, Liu J Zhu H (2017) Zika Virus: An update and its implication for vaccine development. JOJ Immuno Virology 2.
- Smithburn KC, Hughes TP, Burke AW, et al. (1940) A neurotropic virus isolated from the blood of a native of Uganda. Am J Trop Med 20: 471-492.
- Hayes EB, Komar N, Nasci RS, et al. (2005) Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis 11: 1167-1173.
- Guarner J, Shieh WJ, Hunter S, et al. (2004) Clinicopathologic study and laboratory diagnosis of 23 cases with West Nile virus encephalomyelitis. Hum Pathol 35: 983-990.
- Cheeran MC, Hu S, Sheng WS, et al. (2005) Differential responses of human brain cells to West Nile virus infection. J Neurovirol 11: 512-524.
- Hayes C (1989) West nile fever. In: Monath TP, The arboviruses: Epidemiology and ecology. FL: CRC Press, Boca Raton, 59-88.
- Murgue B, Murri S, Triki H, et al. (2001) West Nile in the Mediterranean basin: 1950-2000. Ann N Y Acad Sci 951: 117-126.
- Panthier R, Hannoun C, Oudar J, et al. (1966) Isolement du virus West Nile chez un cheval de Camargue attaint d'encephalomyelite. C R Acad Science Paris 262: 1308-1310.
- Nash D, Mostashari F, Fine A, et al. (2001) The outbreak of West Nile virus infection in the New York City area in 1999. N Engl J Med 344: 1807-1814.
- CDC (2014) West Nile virus and other nationally notifiable arboviral diseases - United States.
- Hayes EB, Gubler DJ (2006) West Nile virus: Epidemiology and clinical features of an emerging epidemic in the United States. Annu Rev Med 57: 181-194.
- http://www.cdc.gov/non-polio-enterovirus/about/EV-D68.html
- Kemmerly SA (2003) Diagnosis and treatment of west nile infections. Ochsner J 5: 16-17.
- Martina BEE, Koraka P, Osterhaus ADME (2010) West Nile Virus: Is a vaccine needed? Curr Opin Investig Drugs 11: 139-146.
- Solomon T, Dung NM, Vaughn DW, et al. (2000) Neurological manifestations of dengue infection. Lancet 355: 1053-1059.
- Gubler DJ (1998) Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11: 480-496.
- Gaunt MW, Sall AA, de Lamballerie X, et al. (2001) Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J Gen Virol 82: 1867-1876.
- Wang EY, Ni HL, Xu RL, et al. (2000) Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol 74: 3227-3234.
- Hayes EB, Gubler DJ (1992) Dengue and dengue hemorrhagic fever. Pediatr Infect Dis J 11: 311-317.
- Halstead SB (1980) Dengue haemorrhagic fever--a public health problem and a field for research. Bull World Health Organ 58: 1-21.
- Yi BT, Zhang ZY (2002) Epidemic and control of dengue fever in China. Chin Public Health 18: 1128-1130.
- Wu JY, Lun ZR, James AA, et al. (2010) Review: Dengue fever in mainland China. Am J Trop Med Hyg 83: 664-671.
- Shepard DS, Coudeville L, Halasa YA, et al. (2011) Economic impact of dengue illness in the Americas. Am J Trop Med Hyg 84: 200-207
- Fredericks AC, Fernandez-Sesma A (2014) The burden of dengue and chikungunya worldwide: implications for the Southern United States and California. Ann Glob Health 80: 466-475.
- Teets FD, Ramgopal MN, Sweeney KD, et al. (2014) Origin of the dengue virus outbreak in Martin County, Florida, USA 2013. Virol Rep 1-2: 2-8.
- CDC (2013) Reemergence of Dengue in Southern Texas.
- Robert Herriman (2017) Dengue fever: 80 cases reported in US and territories in 1st six months of 2017.
- Kiley MP, Bowen ETW, Eddy GA, et al. (1982) Filoviridae: A taxonomic home for marburg and ebola viruses. Intervirology 18: 24-32.
- Leroy EM, Kumulungui B, Pourrut X, et al. (2005) Fruit bats as reservoirs of Ebola virus. Nature 438: 575-576.
- Feldmann H, Slenczka W, Klenk HD (1996) Emerging and reemerging of filoviruses. Arch Virol Suppl 11: 77-100.
- Slenczka W, Klenk HD (2007) Forty years of marburg virus. Journal of Infectious Diseases 196: S131-S135.
- Barrette RW, Metwally SA, Rowland JM, et al. (2009) Discovery of swine as a host for the reston ebolavirus. Science 325: 204-206.
- Jahrling PB, Geisbert TW, Dalgard DW, et al. (1990) Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet 335: 502-505.
- Robert A Lever, Christopher JM Whitty (2016) Ebola virus disease: emergence, outbreak and future directions. British Medical Bulletin 117: 95-106.
- World Health Organization (2016) Zika Strategic Response Framework & Joint Operations Plan.
- Aleksandrowicz P, Marzi A, Biedenkopf N, et al. (2011) Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis 204: S957-S967.
- Nanbo A, Imai M, Watanabe S, et al. (2010) Ebola virus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. Plos Pathog 6: e1001121.
- Zhou NN, Senne DA, Landgraf JS, et al. (1999) Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73: 8851-8856.
- Russell RJ, Kerry PS, Stevens DJ, et al. (2008) Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 105: 17736-17741.
- Russell RJ, Haire LF, Stevens DJ, et al. (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443: 45-49.
- Wright PF, Neumann G, Kawaoka Y (2006) Orthomyxoviruses. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA. Fields Virology. Lippincott Williams & Wilkins, Philadelphia PA, USA, 1691-1740.
- Perroncito E (1878) Epizoozia tifoide nei gallinacei. Annali Accad Agri Torino 21: 87-126.
- Zimmer SM, Burke DS (2009) Historical perspective--Emergence of influenza A (H1N1) viruses. N Engl J Med 361: 279-285.
- Logan WPD, MacKay DG (1951) Development of influenza epidemics. Lancet 1: 264-265.
- Sartwell PE, Long AP (1948) The Army experience with influenza, 1946-1947; epidemiological aspects. Am J Hyg 47: 135-141.
- Salk JE, Suriano PC (1949) Importance of antigenic composition of influenza virus vaccine in protecting against the natural disease; observations during the winter of 1947-1948. Am J Public Health Nations Health 39: 345-355.
- Nelson MI, Viboud C, Simonsen L, et al. (2008) Multiple reassortment events in the evolutionary history of H1N1 influenza a virus since 1918. PLoS pathog 4: e1000012.
- Scholtissek C, Rohde W, Von Hoyningen V, et al. (1978) On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87: 13-20.
- Kilbourne ED (2006) Influenza pandemics of the 20th century. Emerg Infect Dis 12: 9-14.
- Kawaoka Y, Krauss S, Webster RG (1989) Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol 63: 4603-4608.
- Shope RE (1931) Swine influenza: I. experimental transmission and pathology. J Exp Med 54: 349-359.
- Webby RJ, Swenson SL, Krauss SL, et al. (2000) Evolution of swine H3N2 influenza viruses in the United States. J Virol 74: 8243-8251.
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S, et al. (2009) Emergence of a novel swine-origin influenza a (H1N1) virus in humans. N Engl J Med 360: 2605-2615.
- Newman AP, Reisdorf E, Beinemann J, et al. (2008) Human case of swine influenza A (H1N1) triple reassortant virus infection, Wisconsin. Emerg Infectious Dis 14: 1470-1472.
- Shinde V, Bridges CB, Uyeki TM, et al. (2009). Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N Engl J Med 360: 2616-2625.
- Pensaert M, Ottis K, Vandeputte J, et al. (1981) Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ 59: 75-78.
- Centers for Disease Control Prevention (2009) Swine influenza A (H1N1) infection in two children--Southern California, March-April 2009. MMWR Morb Mortal Wkly Rep 58: 400-402.
- George KS (2012) Diagnosis of influenza virus. Methods Mol Biol 865: 53-69.
- South China Morning post, Health and Environment (2017) Mutation may be behind surge in flu cases this summer, Hong Kong experts say.
- Openshaw PJ (2009) Crossing barriers: infections of the lung and the gut. Mucosal Immunol 2:100-102.
- Cherry JD (2004) The chronology of the 2002-2003 SARS mini pandemic. Paediatr Respir Rev 5: 262-269.
- Lee N, Hui D, Wu A, et al. (2003) A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 348: 1986-1994.
- Peiris JS, Lai ST, Poon LL, et al. (2003) Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361: 1319-1325.
- Fleck F (2004) SARS virus returns to China as scientists race to find effective vaccine. Bull World Health Organ 82: 152-153.
- Liang GD, Chen QX, Xu JG, et al. (2004) Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg Infect Dis 10: 1774-1781.
- Guan Y, Zheng BJ, He YQ, et al. (2003) Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 302: 276-278.
- Hui DS, Memish ZA, Zumla A (2014) Severe acute respiratory syndrome vs. the Middle East respiratory syndrome. Curr Opin Pulm Med 20: 233-241.
- Oboho IK, Tomczyk SM, Al-Asmari AM, et al. (2015) 2014 MERS-CoV Outbreak in Jeddah - A Link to Health Care Facilities. The New England Journal of Medicine 372: 846-854.
- Feikin DR, Alraddadi B, Qutub M, et al. (2015) Association of higher MERS-CoV virus load with severe disease and death, Saudi Arabia, 2014. Emerg Infect Dis 21: 2029-2035.
- Munster VJ, Prescott JB, Bushmaker T, et al. (2012) Rapid Nipah virus entry into the central nervous system of hamsters via the olfactory route. Sci Rep 2: 736.
- Halpin K, Mungall BA (2007) Recent progress in henipavirus research. Comp Immunol Microbiol Infect Dis 30: 287-307.
- Lai-Meng, Kaw-Bing CHUA (2007) Lessons from the Nipah virus outbreak in Malaysia. Malays J Pathol 29: 63-67.
- Parashar UD, LM Sunn, F Ong, et al. (2000) Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J Infect Dis, 181: 1755-1759.
- Hsu VP, Hossain MJ, Parashar UD, et al. (2004) Nipah virus encephalitis reemergence, Bangladesh. Emerging infectious diseases 10: 2082-2087.
- Luby SP, Rahman M, Hossain MJ, et al. (2006) Foodborne transmission of nipah virus, Bangladesh. Emerg Infect Dis 12: 1888-1894.
- Pallansch M, Roos R (2007) Enteroviruses: Polioviruses, Coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe DM, Howley PM, Field Virology. (5th edn), Lippincott Williams & Wilkins: Philadelphia, USA, 840-893.
- Adams MJ, Lefkowitz EJ, King AMQ, et al. (2015) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2015). Arch Virol 160: 1837-1850.
- Bessaud M, Razafindratsimandresy R, Nougairede A, et al. (2014) Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS One 9: e90624.
- Schmidt NJ, Lennette EH, Ho HH (1974) An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis 129: 304-309.
- Zhang Y, Tan XJ, Wang HY, et al. (2009) An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J Clin Virol 44: 262-267.
- Zhang Y, Zhu Z, Yang W, et al. (2010) An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol J 7: 94.
- Sun LM, Zheng HY, Zheng HZ, et al. (2011) An enterovirus 71 epidemic in Guangdong Province of China, 2008: epidemiological, clinical, and virogenic manifestations. Jpn J Infect Dis 64:13-18.
- Drake JW (1993) Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A 90: 4171-4175.
- WPRO (2012) Hand, Foot and Mouth Disease (HFMD).
- Departament de Salut of Catalunya (2016) 22 infants ingressats amb afectacions neurològiques agudes per enterovirus.
- Ooi MH, Wong SC, Lewthwaite P, et al. (2010) Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol 9: 1097-1105.
- Centers for Disease Control and Prevention (2011) Clusters of acute respiratory illness associated with human enterovirus 68--Asia, Europe, and United States, 2008-2010. MMWR Morb Mortality wkly rep 60: 1301-1304.
- Khetsuriani N, Lamonte-Fowlkes A, Oberst S, et al. (2006) Enterovirus surveillance-United States, 1970-2005. MMWR Surveillance Summaries 55: 1-20.
- Lauinger IL, Bible JM, Halligan EP, et al. (2012) Lineages, sub-lineages and variants of enterovirus 68 in recent outbreaks. PLoS One 7: e36005.
- Poelman R, Schuffenecker I, Van Leer-Buter C, et al. (2015) European surveillance for enterovirus D68 during the emerging North-American outbreak in 2014. Journal of Clinical Virology 71: 1-9.
- Oermann CM, Schuster JE, Conners GP, et al. (2015) Enterovirus d68. A focused review and clinical highlights from the 2014 U.S. Outbreak. Ann Am Thorac Soc 12: 775-781.
- Edwin J, Reyes Domingo F, Booth T, et al. (2015) Surveillance summary of hospitalized pediatric enterovirus D68 cases in Canada, September 2014. Canada Communicable Disease Report 41(S1).
- Messacar K, Mark J Abzug, Samuel R Dominguez (2016) The Emergence of Enterovirus-D68. Microbiol spectr 4.
- Wylie TN, Wylie KM, Buller RS, et al. (2015) Development and evaluation of an enterovirus D68 Real-Time Reverse Transcriptase PCR Assay. J Clin Microbiol 53: 2641-2647.
- 121. Piralla A, Girello A, Premoli M, et al. (2015) A new real-time reverse transcription-pcr assay for detection of human enterovirus 68 in respiratory samples. J Clin Microbiol 53: 1725-1726.
- Mao Qy, Wang Y, Bian L, et al. (2016) EV71 vaccine, a new tool to control outbreaks of hand, foot, and mouth diseases (HFMD). Expert Rev Vaccines 15: 599-606.
- Morens DM, Fauci AS (2013) Emerging infectious diseases: Threats to human health and global stability. PLoS Pathog 9: e1003467.
- Howard CR, Fletcher NF (2012) Emerging virus diseases: can we ever expect the unexpected? Emerg Microbes Infect 1: e46.
- W Shuaib, H Stanazai, AG Abazid, et al. (2016) Re-emergence of zika virus: A review on pathogenesis, clinical manifestations, diagnosis, treatment, and prevention. Am J Med 129: 879.
- Dick GWA (1952) Zika virus (II). Pathogenicity and physical properties. Transactions of the Royal Society of Tropical Medicine and Hygiene 46: 521-534.
- Ioos S, Mallet HP, Goffart IL, et al. (2014) Current Zika virus epidemiology and recent epidemics. Med Mal Infect 44: 302-307.
- Foy BD, Kobylinski KC, Chilson Foy JL, et al. (2011) Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 17: 880-882.
- https://agriculture.az.gov/pests-pest-control/human-diseases/west-nile
- European center for disease prevention and control (ECDC) (2009) Influenza Virus outbreaks.
Corresponding Author
Hua Zhu, Department of Microbiology, Biochemistry and Molecular Genetics, New Jersey Medical School, Rutgers, USA; College of Life Sciences, Jinan University, Guangzhou, Guangdong 510632, China.
Copyright
© 2018 Jaijyan DK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.