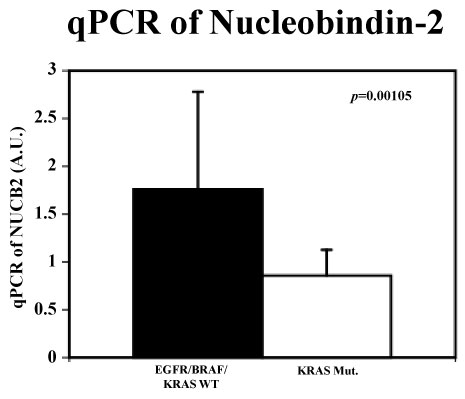
Nucleobindin-2 mRNA level is down regulated in KRAS-mutation lung cancer cell lines compared with EGFR/BRAF/KRAS wild-type lung cancer cell lines
Keywords
Lung cancer, Nucleobindin-2, Nesfatin-1, KRAS, qPCR
Abbreviations
EGF: Epidermal Growth Factor; MEK: Mitogen-Activated Protein Kinase; ERK: Extracellular Signal-Regulated Kinase; GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase
To the editor; Increased Nucleobindin-2 level has been determined as an independent prognostic factor for overall survival of patients [1,2]. However, pathophysiological significance of Nucleobindin-2 in lung cancers remains unclear. In the present study, we examined Nucleobindin-2 mRNA level in various types of human lung cancer cell lines.
Ten of lung cancer cell lines with EGFR/BRAF/KRAS wild-type (H1299, H1819, HCC95, H838, H1437, H661, HCC15, HCC78, H1648, HCC193) and eleven of lung cancer cell lines with KRAS-mutation [KRAS-G12C (H2122, HCC44, H1792, HCC4017, H358), KRAS-G12V (H441), KRAS-G12D (HCC515), KRAS-G12R (H1264, H157), KRAS-G12A (H2009), KRAS-Q61H (H460)] were used [3]. Nucleobindin-2 mRNA level was analyzed by RT-qPCR as previously rescribed [3]. Primers and probes for Nucleobindin-2 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were obtained from Ori Gene Technologies (MK203097; Rockville, MD, USA;) and Applied Biosystems (Assay ID: Hs99999905_m1; Tokyo, Japan;), respectively. To normalize the amount of input cDNA, quantitative analysis was performed using the GAPDH as an internal reference. Relative expression values were computed using the comparative cycle threshold (Ct) method. All data in the figure are presented as mean ± standard deviation and were analyzed using one-way ANOVA to compare the means of all the groups. Turkey-Kramer multiple comparisons method was used to determine statistical differences between the means, with p < 0.05 deemed statistically significant using the InStat 2.00 program.
As shown in Figure 1, we discovered that lung cancer cell lines associated with KRAS-mutations had significantly decreased mRNA level of Nucleobindin-2 compared to lung cancer cells with EGFR/BRAF/KRAS wild-type (p = 0.00105).
KRAS-mutations are almost exclusively detected in lung adenocarcinomas [4]. In the case of non small cell lung cancer (NSCLC), KRAS mutations occur predominantly at codon 12 (> 80%) [4]. The most frequent codon variant is KRAS-G12C mutation (39%) [4]. Other common mutations include KRAS-G12V (18-21%) and KRAS-G12D (17-18%) variants [4]. KRAS-mutant lung cancers have generally been associated with poorer overall survival than KRAS wild-type tumors, particularly in the advanced-stage [5]. In the current study, KRAS-G12C was five of eleven, KRAS-G12V was one of eleven, and KRAS-G12D was one of eleven cell lines. Having those sets of KRAS-mutant lung cancer cell lines, we compared Nucleobindin-2 mRNA levels between EGFR/BRAF/KRAS wild-type and KRAS-mutant lung cancer cell lines. Based on our observation, it was speculated that Nucleobindin-2 was downregulated in KRAS-mutant lung cancer cell lines and this downregulation was nothing to do with KRAS mutation types. Unlike other types of cancers, Nucleobindin-2 seems to play uniqule pathophysiological role on KRAS-mutant lung cancers because of unique downregulation instead of up regulation. When the relationship of Nucleobindin-2 levels and clinical significances are evaluated in lung cancer patients, reserachers need to pay attention about the presence or absence of KRAS mutation.
Acknowledgements
Lung cancer cell lines (H1299, H1819, HCC95, H838, H1437, H661, HCC15, HCC78, H1648, HCC193, H2122, HCC44, H1792, HCC4017, H441, H358, HCC515, H1264, H157, H2009, H460) used in this study were generously provided by Drs John D. Minna and Adi F. Gazdar of the University of Texas Southwestern Medical Center at Dallas.
Disclosure
None of the authors has any potential conflicts of interest associated with this case report.
Authors' Contributions
All authors had active participation in preparation of the manuscript. All authors read and approved the final manuscript.
Ethical Approval and Consent
The Ethics Committee of Gunma University Hospital approved this study.
Consent to Publish
All participants understand that the information will be published anonymously, but that full anonymity cannot be guaranteed. We understand that the text and any pictures or videos published in the article will be freely available on the internet and may be seen by the general public. Pictures, videos, and text may also appear on other websites or in print, may be translated into other languages or used for commercial purposes.
Competing Interests
We declare that we have no significant competing financial, professional, or personal interests that may have influenced the performance or presentation of the work described in this manuscript.
References
- Xie J, Chen L, Chen W (2018) High NUCB2 expression level is associated with metastasis and may promote tumor progression in colorectal cancer. Oncol Lett 15: 9188-9194.
- Altan B, Kaira K, Okada S, et al. (2017) High expression of nucleobindin 2 is associated with poor prognosis in gastric cancer. Tumour Biol 39: 1010428317703817.
- Sunaga N, Kaira K, Tomizawa Y, et al. (2014) Clinicopathological and prognostic significance of interleukin-8 expression and its relationship to KRAS mutation in lung adenocarcinoma. Br J Cancer 110: 2047-2053.
- Ferrer I, Zugazagoitia J, Herbertz S, et al. (2018) KRAS-Mutant non-small cell lung cancer: From biology to therapy. Lung Cancer 124: 53-64.
- Mascaux C, Iannino N, Martin B, et al. (2005) The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br J Cancer 92: 131-139.
Corresponding Author
Shuichi Okada, Department of Medicine and Molecular Science, Gunma University Graduate School of Medicine, 3-39-15 Showa-machi, Maebashi, Gunma, 371-8511, Japan.
Copyright
© 2019 Sunaga N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.