A Review on High Throughput Phenotyping for Vegetable Crops
Abstract
Conventional phenotyping approaches for vegetable crops such as Solanaceae, Bulb, and Root crops have contributed significantly to the development of numerous varieties. Despite this, traditional phenotyping procedures are insufficient because of the longer time required to produce a variety, poor genetic gain, environmental influences, and other externalities that impact phenotype-based selection. A novel recent approach of high throughput phenotyping (HTP) is regarded a potential tool for addressing the problems of traditional phenotyping. The advent of sensor, computer vision, automation, and sophisticated machine learning technologies sparked the creation of high-throughput phenotyping technology in the prior decade. HTP platforms are being used to conduct non-destructive evaluations of the whole plant system in a variety of crops. HTP provides precise measurements and suggests the collection of high-quality and accurate data, which is required for standardizing phenotyping for genetic dissection and genomic assisted breeding techniques such as genome-wide association studies (GWAS), linkage mapping, marker-assisted selection (MAS), and genomic selection (GS). The rest of this chapter examines the application of high-throughput phenotyping tools in genomic-assisted breeding for vegetable crops.
Keywords
Vegetables, High throughput phenotyping, Genomic assisted breeding
Introduction
The availability of resources for farmers is minimal, making crop management strategies that maximize crop output difficult to implement [1]. Agriculture systems that are over managed may be detrimental to a sustainable agricultural system [2]. While improved genetics, management, and environmental adaptations contributed to the increased production of major commodity crops, quantifying their relative contributions is difficult due to the environment's complex interactions and dynamic nature and management practices [3]. Crop managers face significant challenges in maintaining a consistent and high-yielding crop production level in an unexpected climate, as crop management tactics rely heavily on prior practices [4]. The discrepancy between potential and actual agricultural yields may be significant for certain crops [5]. It is suggested in studies that yield may be enhanced when both best-adapted variety and agronomic practices are applied in the field [5].
However, the improvement of the genetic structure of plants increased the complex trait like yield [6]. To maximize agricultural productivity, crop management practices must address a variety of practical constraints [7]. Moreover, the advancements in breeding technology continue to promote yield gains in staple crops globally [8,9].
Conventional phenotyping techniques are prohibitively expensive, time-consuming, slow, and frequently harmful, and they only allow for the analysis of a few variables at a time [10]. However, traditional breeding operations are being transformed into more efficient contemporary breeding programs by incorporating emerging technologies, most notably high-throughput phenotyping [11].
A new technology, non-destructive phenotyping, adds a new dimension to the data collection process by increasing the precision, speed, and analysis of captured data [12]. According to scientists, agricultural productivity is expected to increase significantly shortly due to genetic enhancement enabled by high-throughput phenotyping technologies [13]. Sensing technology, data processing, and analysis advancements have significantly improved field and crop management tactics [14]. Apart from focusing on a variety of traits that indicate the water content, chlorophyll content, biomass, and growth potential of a plant [15].
However, the development of novel vegetable varieties and target environments poses significant challenges in terms of high-throughput and precision phenotyping, modeling, and collaboration with vegetable breeders [16]. Accurate phenotyping was required for several aspects like physiological, morphological, structural, biochemical and molecular characteristics to develop high-yielding vegetable cultivars that were more resistant to biotic and abiotic stresses [17,18]. As a result, breeders can conduct multiple trials under various growth conditions and with a variety of lines to map populations, breed populations, mutant populations, and the germplasm pool [19]. The remainder of this chapter discusses how high-throughput phenotyping technologies can be used to optimize breeding operations in genomic assisted breeding for vegetable yield gains.
What is High-Throughput Vegetable Phenotyping?
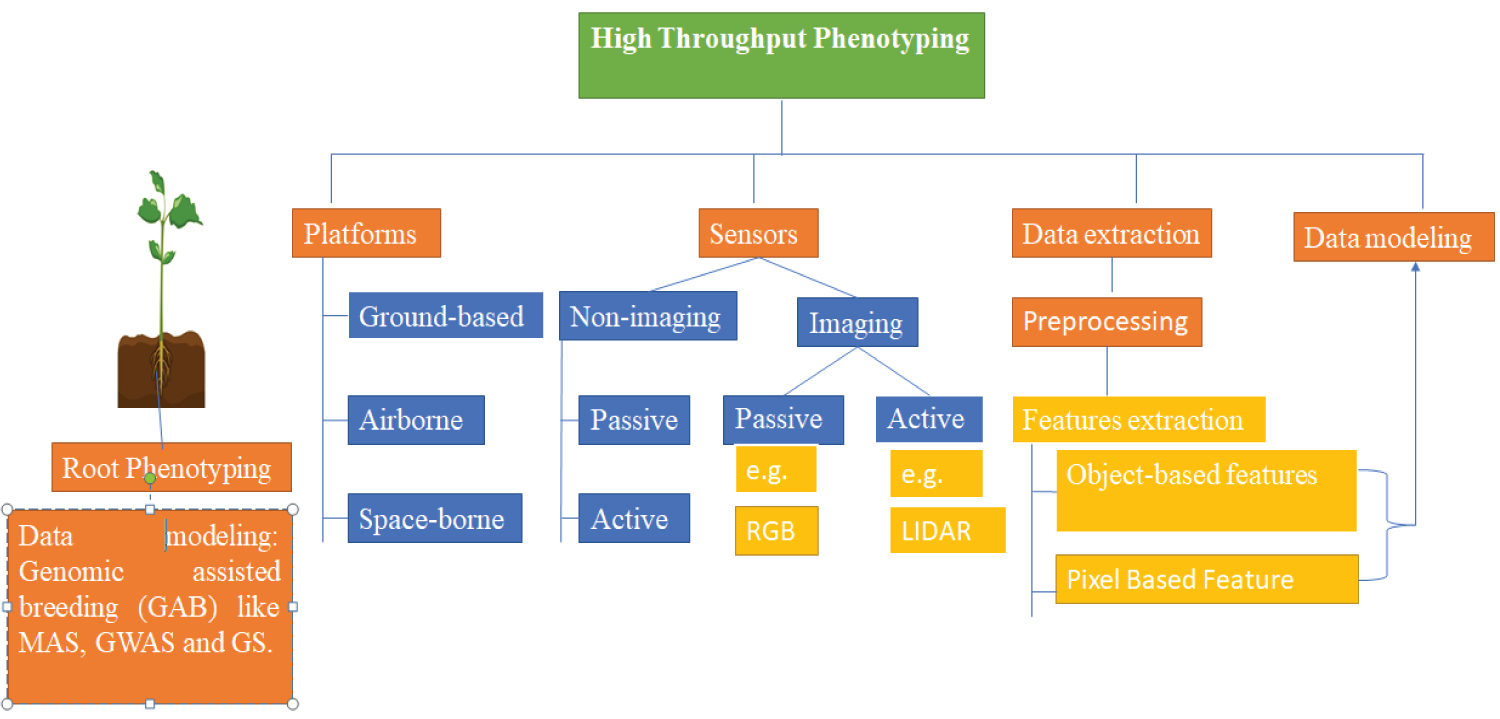
One hundred years ago (Johannsen 1903, 1911), The term "phenotype" was coined as a counterpoint to the concept of "genotypes" [20], and refers to a collection of methodologies and processes for accurately assessing plant growth, architecture, and composition at various sizes [21,22]. Historically, plant breeders have analyzed hundreds to thousands of plant phenotypes using visual observations, manual tools and among other techniques [23-25]. Information on the tools for HTP in vegetables is provided in Figure 1.
A fully functional HTP system is composed of supporting hardware (sensors and platforms) and a computing component that communicates with one another (data process and analytics) [26]. The research will analyze and integrate a variety of advanced imaging techniques commonly used in computed tomography (CT), into HTP systems in this rapidly growing market [27]. While the industrial sector is driving sensor technology advancements, efforts are being made to incorporate them into agricultural high-throughput systems (AHTP) [28].
The data processing and the analytic system is the most critical component of an HTP system. The current generation of HTP systems, particularly those with high-resolution imaging capabilities, can collect multidimensional data on crops from a large number of people [29]. On the other hand, researchers will quickly discover that they are capable of being overwhelmed by massive amounts of data [30]. In conjunction with ongoing community initiatives, HTP technology has the potential to play a critical role in resolving the breeder's dilemma and expediting the development of new crop varieties with advanced traits [31].
A uniform set of criteria for assessing agricultural qualities in multiple dimensions can be achieved using equipment such as spectrum reflectance, photogrammetry, and computer vision [32]. Timely and accurate measurements of agricultural characteristics High-throughput phenotyping devices enable breeding programs to increase their capacity to manage a larger breeding population while maintaining the same level of selection intensity [12]. For example, HTP platforms based on unmanned aerial systems (UAS) could be used to rapidly scan breeding grounds [33]. Advanced sensors capture information about the crop that the human eye or senses are unable to see or perceive [34].
Advanced data analytics and artificial intelligence models extract previously undiscovered information from human and sensor data, and they hold enormous promise for identifying novel agricultural characteristics [35]. The additional elements can be used to characterize plant performance during a particular developmental stage (for example, emerging, blooming, or harvesting) or to assess crop dynamic responses to environmental changes over the course of a growing season [36]. Along with increasing the amount of data available for assessing minute genetic differences between genotypes, unique crop characteristics have the potential to increase genetic diversity within the crop population [36].
Nowadays, genetic studies on QTL mapping and genome-wide association studies (GWAS) may be used to identify critical genetic variables underlying or associated with yield increase using HTP-based phenotypes [37]. By utilizing marker-assisted selection (MAS) during breeding, it is possible to improve the incorporation of genetic characteristics associated with desirable agricultural characteristics into the existing vegetable germplasm [38]. Integrating breeding populations enables more precise selection, shorter breeding cycles, and increased genetic gain [39]. Large-scale phenotyping enables the collection of massive amounts of agricultural data with high spatiotemporal resolution and the identification of novel crop characteristics [40]. This technique enables the integration of crop and environmental data, as well as management data [41]. Prescriptive phenotyping allow the breeders to develop crop qualities in response to the breeder and consumer requests [42].
High-Throughput Phenotyping (HTP) Platforms for Vegetable Crops
High throughput phenotyping is a non-destructive technology that creates a good way for measuring the plant phenotype under laboratory and field circumstances [43]. Using sophisticated automation and robotics, imaging (2D and 3D) methods, innovative sensors, hardware and software, these systems monitors a range of plant growth and development aspects [44]. HTP is based on real-time monitoring of plant growth and development in smart glasshouses and physiological and biochemical reactions [45]. Along with plant growth rates and biomass accumulation, the visible imaging system quantifies a range of characteristics, such as canopy architecture and phenology [46]. On the other hand, a hyperspectral imaging system can identify internal properties such as sugar, starch, protein, and moisture content, as well as a range of factors associated with stress [47]. Multiple pictures at varying time intervals and wavelengths are acquired by HTP devices to create data for software-based analysis [48-52].
High-Throughput Phenotyping in Genomic Assisted Breeding
The advanced technology of high throughput phenotyping has been successfully used for rapid evaluation of plants traits in glass houses and controlled conditions [12]. Both the academic and industrial sectors have attempted to develop high-throughput screening (HTS) technologies to adapt various crops including vegetables for a variety of breeding purposes [46]. Jansen and colleagues (2009) developed the GROWSCREEN FLUORO to assess stress tolerance in rosette plants using leaf growth and chlorophyll fluorescence characteristics [53]. Flood, et al. (2016) developed the Phenovator, which can screen over 1000 Arabidopsis plants for photosynthesis, growth, and multispectral reflectance multiple times per day [54,55]. These studies aimed to decipher agriculture's chronological evolution and assess crop's genetic responses [56].
In vegetable studies, image features are frequently used to replace manual measurements and increase data collection efficiency (phenotyping), or in conjunction with genomic analyses such as quantitative trait loci (QTLs) and genome wide association studies (GWAS) mapping to evaluate genetic variation in crops [57,58] or to predict crop performance [58]. Several quantitative traits are identified in vegetables crops.
High-Throughput Phenotyping under Controlled Condition
A regulated environment is frequently defined in plant science as an enclosed enclosure in which certain environmental variables such as light condition, temperature, humidity temperature and CO2 level are controlled and monitored [59]. Greenhouses, growth chambers, temperature chambers, and nursery rooms are only a few facilities often used in plant research to investigate plant responses to controlled environmental conditions [60]. In controlled environments, plant phenotyping systems are made of sensors, automated control systems, data processing, management systems, and computer software that all work in concert to provide results. The controlled environment is smaller (diameters) and more equipped than natural habitats, which simplifies the deployment of automated phenotyping devices much more than in the wild [61]. These systems collect data on agricultural attributes in a high-throughput manner through the use of sensors, automation, and control systems [62]. The current state of high-throughput plant phenotyping systems in controlled environments was discussed and the sensors used to assess plant characteristics in such systems [63].
Root Phenotyping
Although the root system dictates the positioning of roots in the soil, little is known about the roots of plants when they are not in the soil [64]. It is critical to understand the anatomical properties of roots in order to appreciate water transport, nutrient absorption, root carbon costs, and root interactions with microorganisms such as mycorrhizal fungi [65]. Finally, the most unclear root phenes are those that are reliant on physiological and flux-related processes. When it comes to root research, they are seldom quantified and have received far less attention in "high-throughput" settings than in traditional ones [66]. According to current thought, the physiological phenes of roots represent a vast and unexplored frontier in root research. Roots are notoriously difficult to analyze [67]. As a result of this difficulty, the genetic and functional foundations of root phenes are less established than those of aboveground phenes [68,69].
To close this "phenotyping gap," a shift away from traditional phenotyping toward image-based phenotyping has occurred, which enables relatively high throughput while maintaining root measurement accuracy [70]. Numerous platforms make use of two-dimensional imaging via cameras and propagate plants via seedlings on agar plates, germination paper, or fabric cloth in bins [71,72]. Additionally, readers are encouraged to peruse this manual (Figure 1). Even though controlling environmental factors is advantageous for characterizing root phenotypes, this chapter focuses on strategies applicable to field-grown plants [73]. The integration of root phenes and functional phenomics will need the phenotyping of several root phenes at the same time. It is anticipated that standard approaches will be tested in the field, which will address the root cause of the "phenotyping gap." We must dig deeper and harder to fully realize the promise of roots for agricultural revolutionization [73].
Conclusion
In summary, agricultural HTP technology has the ability to solve the breeder's equation for maximum genetic gain by increasing the intensity and precision of selection, improving the detection of genetic variations, and decreasing breeding cycles. Crop HTP technology is a multidisciplinary and comprehensive approach that integrates research in agronomy, information science, engineering sciences, and biology. Additionally, it leverages cutting-edge computer and artificial intelligence technologies to provide a more comprehensive solution. Numerous advanced data analysis techniques (e.g., machine learning, deep learning) are being used to examine the different phenotypic data available for crops and develop predictive and prescriptive models for crop phenotyping in a highly automated, multi-dimensional, big-data environment. This section will provide the most current information on HTP technology and its applications in plant breeding, genetics, genomics assisted breeding, and some case studies to assist future researchers in developing and improving HTP technology.
References
- Shah F, Wu W (2019) Soil and crop management strategies to ensure higher crop productivity within sustainable environments. Sustainability 11: 1485.
- Castillo-Díaz FJ, Belmonte-Ureña LJ, Camacho-Ferre F, et al. (2021) The management of agriculture plastic waste in the framework of circular economy. Case of the Almeria Greenhouse (Spain). Int J Environ Res Public Health 18: 12042.
- Chapman SC, Chakraborty S, Dreccer MF, et al. (2012) Plant adaptation to climate change-opportunities and priorities in breeding. Crop and Pasture Science 63: 251-268.
- Pearce D, Dora M, Wesana J, et al. (2018) Determining factors driving sustainable performance through the application of lean management practices in horticultural primary production. Journal of Cleaner Production 203: 400-417.
- Beres BL, Hatfield JL, Kirkegaard JA, et al. (2020) Toward a better understanding of genotype × environment × management interactions-A global wheat initiative agronomic research strategy. Front Plant Sci 11: 828.
- Bailey-Serres J, Parker JE, Ainsworth EA, et al. (2019) Genetic strategies for improving crop yields. Nature 575: 109-118.
- Quisumbing AR, Pandolfelli L (2010) Promising approaches to address the needs of poor female farmers: Resources, constraints, and interventions. World Development 38: 581-592.
- Andorf C, Beavis WD, Hufford M, et al. (2019) Technological advances in maize breeding: Past, present and future. Theoretical and Applied Genetics 132: 817-849.
- Schroeder P, Anggraeni K, Weber U (2019) The relevance of circular economy practices to the sustainable development goals. Journal of Industrial Ecology 23: 77-95.
- Hein NT, Ciampitti IA, Jagadish SV (2021) Bottlenecks and opportunities in field-based high-throughput phenotyping for heat and drought stress. J Exp Bot 72: 5102-5116.
- Crossa J, Fritsche-Neto R, Montesinos-Lopez OA, et al. (2021) The modern plant breeding triangle: Optimizing the use of genomics, phenomics, and enviromics data. Front Plant Sci 12: 651480.
- Yang W, Feng H, Zhang X, et al. (2020) Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol Plant 13: 187-214.
- Reynolds M, Chapman S, Crespo-Herrera L, et al. (2020) Breeder friendly phenotyping. Plant Sci 295: 110396.
- Zhou J, Nguyen HT (2021) Solve the breeder’s equation using high-throughput crop phenotyping technology. In: High-Throughput crop phenotyping. Springer, Cham, 1-11.
- Araus JL, Kefauver SC, Zaman-Allah M, et al. (2018) Translating high-throughput phenotyping into genetic gain. Trends plant sci 23: 451-466.
- Yost MA, Kitchen NR, Sudduth KA, et al. (2019) A long-term precision agriculture system sustains grain profitability. Precision Agriculture 20: 1177-1198.
- Westerband AC, Funk JL, Barton KE (2021) Intraspecific trait variation in plants: A renewed focus on its role in ecological processes. Ann Bot 127: 397-410.
- Tripodi P, Massa D, Venezia A, et al. (2018) Sensing technologies for precision phenotyping in vegetable crops: current status and future challenges. Agronomy 8: 57.
- Khadka K, Earl HJ, Raizada MN, et al. (2020) A physio-morphological trait-based approach for breeding drought tolerant wheat. Front Plant Sci 1: 715.
- Ahmar S, Gill RA, Jung KH, et al. (2020) Conventional and molecular techniques from simple breeding to speed breeding in crop plants: Recent advances and future outlook. Int J Mol Sci 21: 2590.
- Thudi M, Palakurthi R, Schnable JC, et al. (2021) Genomic resources in plant breeding for sustainable agriculture. Journal of Plant Physiology 257: 153351.
- Engels JM, Ebert AW (2021) A critical review of the current global ex situ conservation system for plant agrobiodiversity. I. History of the development of the global system in the context of the political/legal framework and its major conservation components. Plants 10: 1557.
- Walter A, Liebisch F, Hund A (2015) Plant phenotyping: From bean weighing to image analysis. Plant Methods 11: 14.
- Costa C, Schurr U, Loreto F, et al. (2019) Plant phenotyping research trends, a science mapping approach. Front Plant Sci 9: 1933.
- Stanschewski CS, Rey E, Fiene G, et al. (2021) Quinoa phenotyping methodologies: An international consensus. Plants 10: 1759.
- Omari MK, Lee J, Faqeerzada MA, et al. (2020) Digital image-based plant phenotyping: A review. Korean Journal of Agricultural Science 47: 119-130.
- Zhang Y, Zhang N (2018) Imaging technologies for plant high-throughput phenotyping: A review. Frontiers of Agricultural Science and Engineering 5: 406-419.
- Piovesan A, Vancauwenberghe V, Van De Looverbosch T, et al. (2021) X-ray computed tomography for 3D plant imaging. Trends in Plant Science 26: 1171-1185.
- Koltes JE, Cole JB, Clemmens R, et al. (2019) A vision for development and utilization of high-throughput phenotyping and big data analytics in livestock. Front Genet 10: 1197.
- Jung J, Maeda M, Chang A, et al. (2021) The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr Opin Biotechnol 70: 15-22.
- Liu Y, Ma X, Shu L, et al. (2020) From Industry 4.0 to Agriculture 4.0: Current status, enabling technologies, and research challenges. IEEE Transactions on Industrial Informatics 17: 4322-4334.
- Louwaars N, Jochemsen H (2021) An ethical and societal analysis for biotechnological methods in plant breeding. Agronomy 11: 1183.
- Oliveira RA, Näsi R, Niemeläinen O, et al. (2020) Machine learning estimators for the quantity and quality of grass swards used for silage production using drone-based imaging spectrometry and photogrammetry. Remote Sensing of Environment 246: 111830.
- Gao J, Westergaard JC, Alexandersson E (2021) Computer vision and less complex image analyses to monitor potato traits in fields. In: David Dobnik, Kristina Gruden, Živa Ramšak, et al. Solanum tuberosum. Humana, New York, NY, 273-299.
- Collins C, Dennehy D, Conboy K, et al. (2021) Artificial intelligence in information systems research: A systematic literature review and research agenda. International Journal of Information Management 60: 102383.
- Furbank RT, Tester M (2011) Phenomics-technologies to relieve the phenotyping bottleneck. Trends in plant science 16: 635-644.
- Rufo R, López A, Lopes MS, et al. (2021) Identification of QTL hotspots affecting agronomic traits and high-throughput vegetation indices in rainfed wheat. bioRxiv.
- Kamboj D, Kumar S, Mishra CN, et al. (2020) Marker assisted breeding in cereals: Progress made and challenges in India. Journal of Cereal Research 12: 85-102.
- Cobb JN, Juma RU, Biswas PS, et al. (2019) Enhancing the rate of genetic gain in public-sector plant breeding programs: lessons from the breeder’s equation. Theoretical and Applied Genetics 132: 627-645.
- Kar S, Garin V, Kholová J, et al. (2020) SpaTemHTP: a data analysis pipeline for efficient processing and utilization of temporal high-throughput phenotyping data. Front Plant Sci 11: 552509.
- Leggieri PA, Liu Y, Hayes M, et al. (2021) Integrating systems and synthetic biology to understand and engineer microbiomes. Annu Rev Biomed Eng 23: 169-201.
- Shook J, Gangopadhyay T, Wu L, et al. (2021) Crop yield prediction integrating genotype and weather variables using deep learning. PLoS One 16: 0252402.
- Chapman SC, Merz T, Chan A, et al. (2014) Pheno-copter: A low-altitude, autonomous remote-sensing robotic helicopter for high-throughput field-based phenotyping. Agronomy 4: 279-301.
- Li L, Zhang Q, Huang D (2014) A review of imaging techniques for plant phenotyping. Sensors 14: 20078-20111.
- Mahlein AK (2016) Plant disease detection by imaging sensors-parallels and specific demands for precision agriculture and plant phenotyping. Plant Disease 100: 241-251.
- Furbank RT, Jimenez-Berni JA, George-Jaeggli B, et al. (2019) Field crop phenomics: Enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol 223: 1714-1727.
- Slater AT, Cogan NO, Rodoni BC, et al. (2017) Breeding differently-the digital revolution: high-throughput phenotyping and genotyping. Potato Research 60: 337-352.
- Schapiro D, Yapp C, Sokolov A, et al. (2022) MITI Minimum Information guidelines for highly multiplexed tissue images. Nat Methods 19: 262-267.
- Pask A, Pietragalla J (2012) Leaf area, green crop area and senescence. In: Pask A, Pietragalla J, Mullan D, et al. Physiological breeding II: A field guide to wheat phenotyping. CIMMYT, Mexico, 58-62.
- Busemeyer L, Mentrup D, Möller K, et al. (2013) BreedVision-A multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors 13: 2830-2847.
- Yol E, Toker C, Uzun B (2015) Traits for phenotyping. In: Phenomics in crop plants: trends, options and limitations. Springer, New Delhi, India, 11-26.
- Zhou J, Ye H, Nguyen HT (2021) High-Throughput Crop Phenotyping Systems for Controlled Environments. In High-Throughput Crop Phenotyping. Springer, Cham 183-208.
- York LM (2021) Phenotyping root system architecture, anatomy, and physiology to understand soil foraging. In: High-Throughput crop phenotyping. Springer, Cham 209-221.
- Bates K (2020) Tools for behavioral phenotyping of C. Elegans.
- Chawade A, Van Ham J, Blomquist H, et al. (2019) High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 9: 258.
- Jansen M, Gilmer F, Biskup B, et al. (2009) Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36: 902-914.
- Zhou J, Fu X, Zhou S, et al. (2019) Automated segmentation of soybean plants from 3D point cloud using machine learning. Computers and Electronics in Agriculture 162: 143-153.
- Dornbusch T, Lorrain S, Kuznetsov D, et al. (2012) Measuring the diurnal pattern of leaf hyponasty and growth in Arabidopsis-A novel phenotyping approach using laser scanning. Funct Plant Biol 39: 860-869.
- Nephali L, Piater LA, Dubery IA, et al. (2020) Biostimulants for plant growth and mitigation of abiotic stresses: A metabolomics perspective. Metabolites 10: 505.
- Chang F, Lv W, Lv P, et al. (2021) Exploring genetic architecture for pod-related traits in soybean using image-based phenotyping. Molecular Breeding 41: 1-21.
- Atieno J, Li Y, Langridge P, et al. (2017) Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci Rep 7: 1300.
- Roy J, Rineau F, De Boeck HJ, et al. (2021) Ecotrons: powerful and versatile ecosystem analysers for ecology, agronomy and environmental science. Glob Chang Biol 27: 1387-1407.
- Paradiso R, Proietti S (2021) Light-Quality manipulation to control plant growth and photomorphogenesis in greenhouse horticulture: The state of the art and the opportunities of modern LED systems. Journal of Plant Growth Regulation 41: 742-780.
- Fan J, Zhang Y, Wen W, et al. (2021) The future of Internet of Things in agriculture: Plant high-throughput phenotypic platform. Journal of Cleaner Production 280: 123651.
- Rouphael Y, Spíchal L, Panzarová K, et al. (2018) High-throughput plant phenotyping for developing novel biostimulants: From lab to field or from field to lab? Front Plant Sci 9: 1197.
- Hou LH, Gao W, Weng ZH, et al. (2021) Use of X-ray tomography for examining root architecture in soils. Geoderma 405: 115405.
- Lynch JP, Strock CF, Schneider HM, et al. (2021) Root anatomy and soil resource capture. Plant and Soil 466: 21-63.
- Wang T, Rostamza M, Song Z, et al. (2019) SegRoot: A high throughput segmentation method for root image analysis. Computers and Electronics in Agriculture 162: 845-854.
- Eshel A, Beeckman T (2013) Plant roots: The hidden half. (4 th edn), CRC Press, Florid.
- Mir RR, Reynolds M, Pinto F, et al. (2019) High-throughput phenotyping for crop improvement in the genomics era. Plant Science 282: 60-72.
- Yasrab R, Atkinson JA, Wells DM, et al. (2019) RootNav 2.0: Deep learning for automatic navigation of complex plant root architectures. Gigascience 8: 123.
- Tracy SR, Nagel KA, Postma JA, et al. (2020) Crop improvement from phenotyping roots: Highlights reveal expanding opportunities. Trends Plant Sci 25: 105-118.
- Paul K, Sorrentino M, Lucini L, et al. (2019) Understanding the biostimulant action of vegetal-derived protein hydrolysates by high-throughput plant phenotyping and metabolomics: A case study on tomato. Front Plant Sci 10: 47.
Corresponding Author
Prashant Kaushik, Kikugawa Research Station, Yokohama Ueki, 2265, Kamo, Kikugawa City, Shizuoka 439-0031, Japan; Instituto de Conservación y Mejora de la Agrodiversidad Valenciana, Universitat Politècnica de València, 46022 Valencia, Spain.
Copyright
© 2023 Kumar A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.