Effects of Co-occurring Genomic Alterations on Survival in Patients with KRAS-Mutant Pancreatic Cancer
Abstract
Pancreatic Cancer is one of the most malignant cancers with a high mortality rate. Although the effects of mutant-KRAS gene have been highly studied in the past, the effects of genes co-mutated with mutant KRAS lack a clear explanation. The study reveals that co-mutating genes also hold a significant role in inducing pancreatic cancer in patients, specifically divided into three classes of groups: Cell adhesion, cell growth/maintenance, and cell communication. The KRAS mutation may co-occur with other gene-mutations, resulting in the effects of mutant-KRAS on patient survival to be compounded by the presence of co-occurring genomic mutations. Our study reveals that amongst the categories of co-mutated genes, cell communication is found to hold the most severe effect on patient survivability.
Introduction
Pancreatic cancer, including Pancreatic Adenocarcinoma and PNET (Pancreatic Neuroendocrine Tumor), is known to be one of the most common and deadly cancers [1] in our society. Numerous studies [2] have shown a strong relationship between mutations in the KRAS gene, a gene that codes for a protein that regulates the propagation of growth factors and cell signaling molecules, and the development of pancreatic cancer (Figure 1). Acting as a molecular on/off switch in guanosine triphosphate (GTP) activity, KRAS binds to GTP until deactivation caused by the conversion of GTP into guanosine diphosphate (GDP). Mutant forms of the KRAS gene induces an uninterrupted activation of the protein despite the lack of extracellular signals; thus, the excessive activation of KRAS causes the formation of tumor in the pancreas.
While the significance of KRAS and its mutant form in promoting pancreatic cancer is heavily studied, there have been few studies analyzing the role of genes that co-occur with mutant-KRAS. Although mutant KRAS has been considered mutually exclusive driver mutations for a long time, there is now growing evidence that KRAS-mutated pancreatic cancer represents a genetically heterogeneous subgroup, specifically divided into categories of molecules responsible for cell adhesion, cell communication, and cell growth/maintenance. The following study analyzes the relationship between mutations in the co-occurring genes and the development of pancreatic cancer. We then explore the effects of co-mutations on patient survivability and on various other pathways.
Discussion
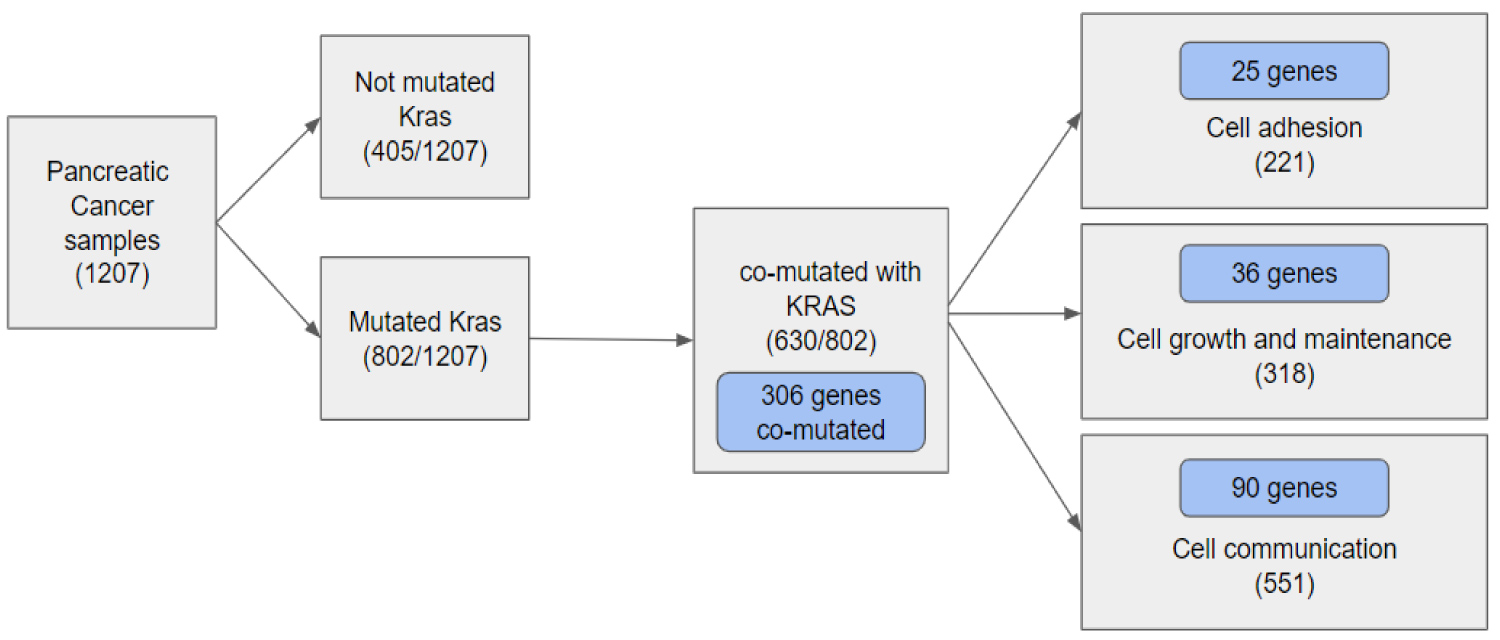
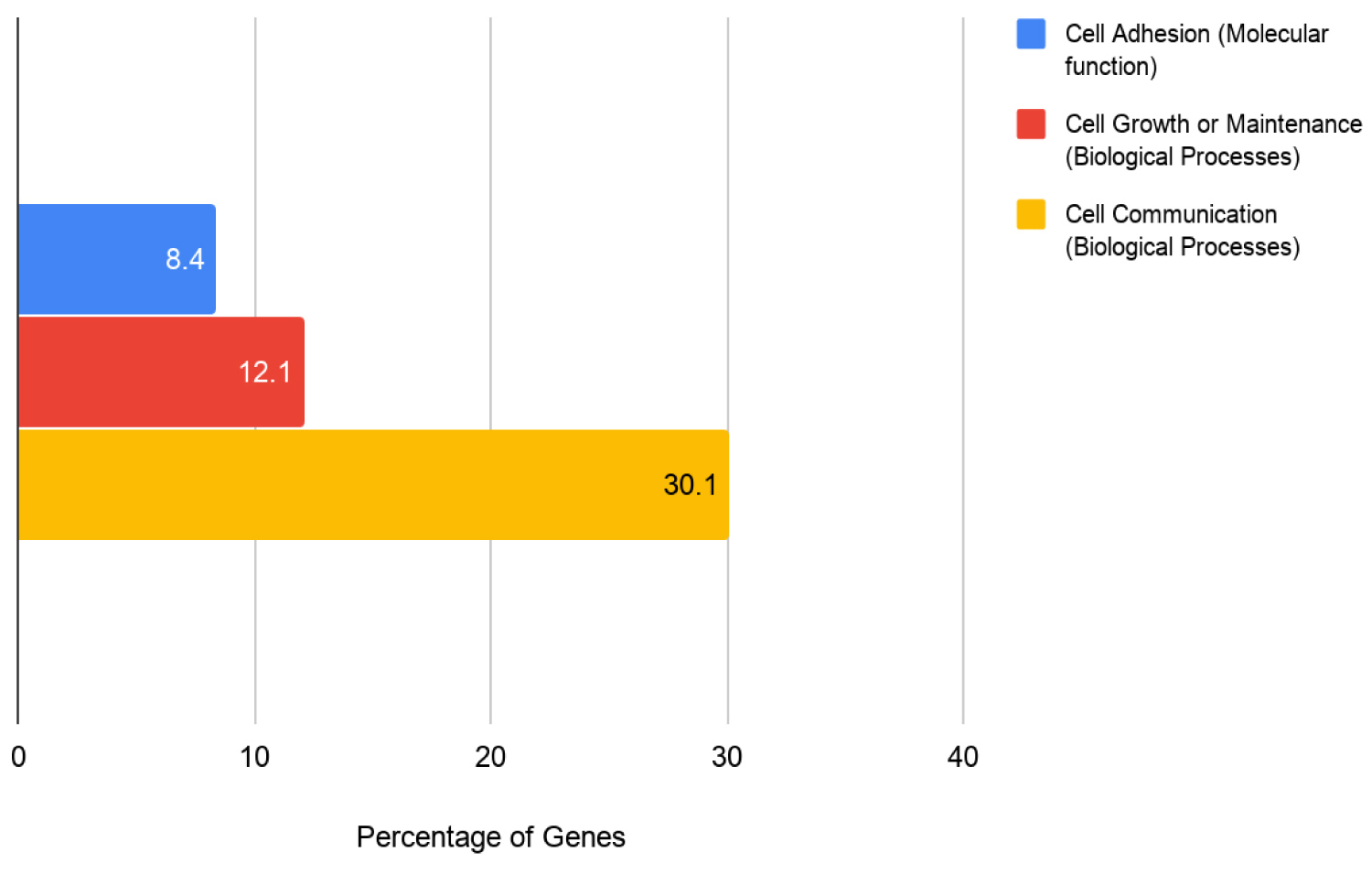
The study found the various effects of co-occurring genomic alterations on survival in patients with KRAS-Mutant Pancreatic cancer. The study analyzes the genetic heterogeneity with respect to cancer-related co-mutations and their correlation with KRAS mutation. As the pattern of co-occurring mutations may describe different biological subsets of patients with KRAS-mutant pancreatic cancer, we explored the effects of co-occurring mutations on patient survival rates. Our results, based on 1207 patient samples from 10 different studies, show that a total of 306 co-occurring mutations occur in patients with KRAS-mutant pancreatic cancer. Further analysis was performed by utilizing the FunRich tool, categorizing the 306 co-occurring genes into three biological/molecular groups to analyze the distinct effects each group holds to the survivability of pancreatic cancer patients (Figure 2, Figure 3 and Figure 4).
Cell communication
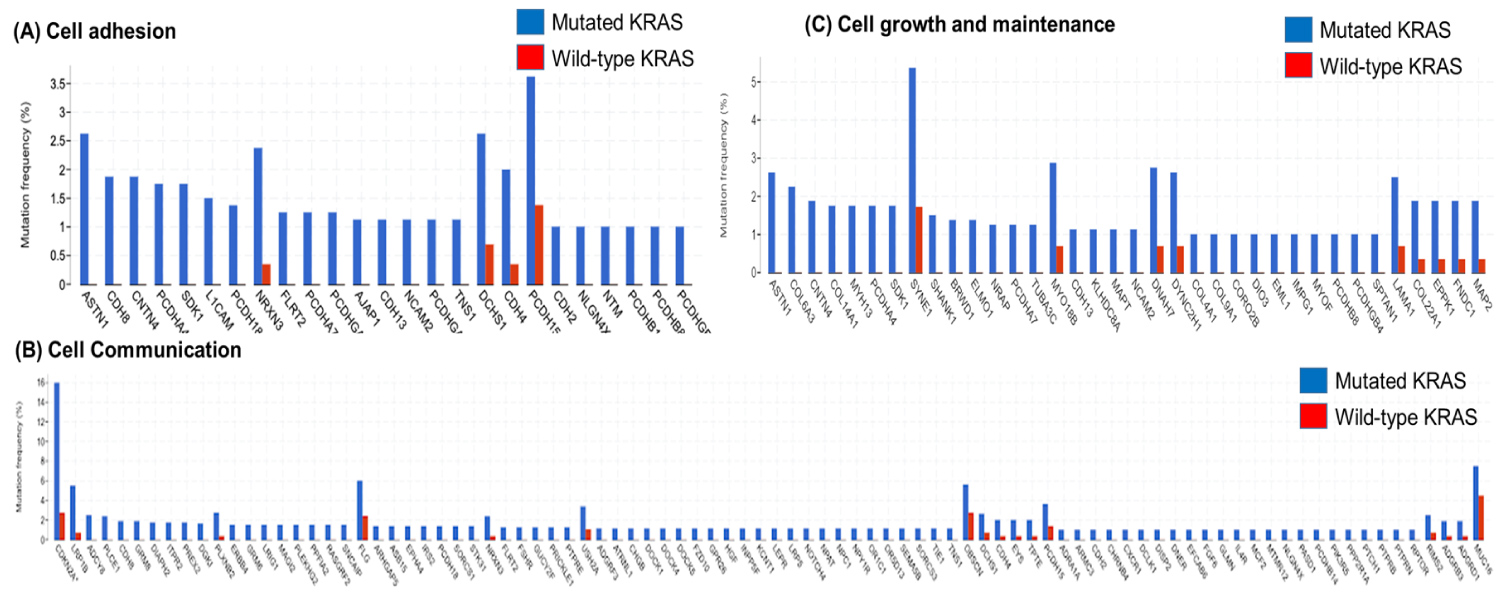
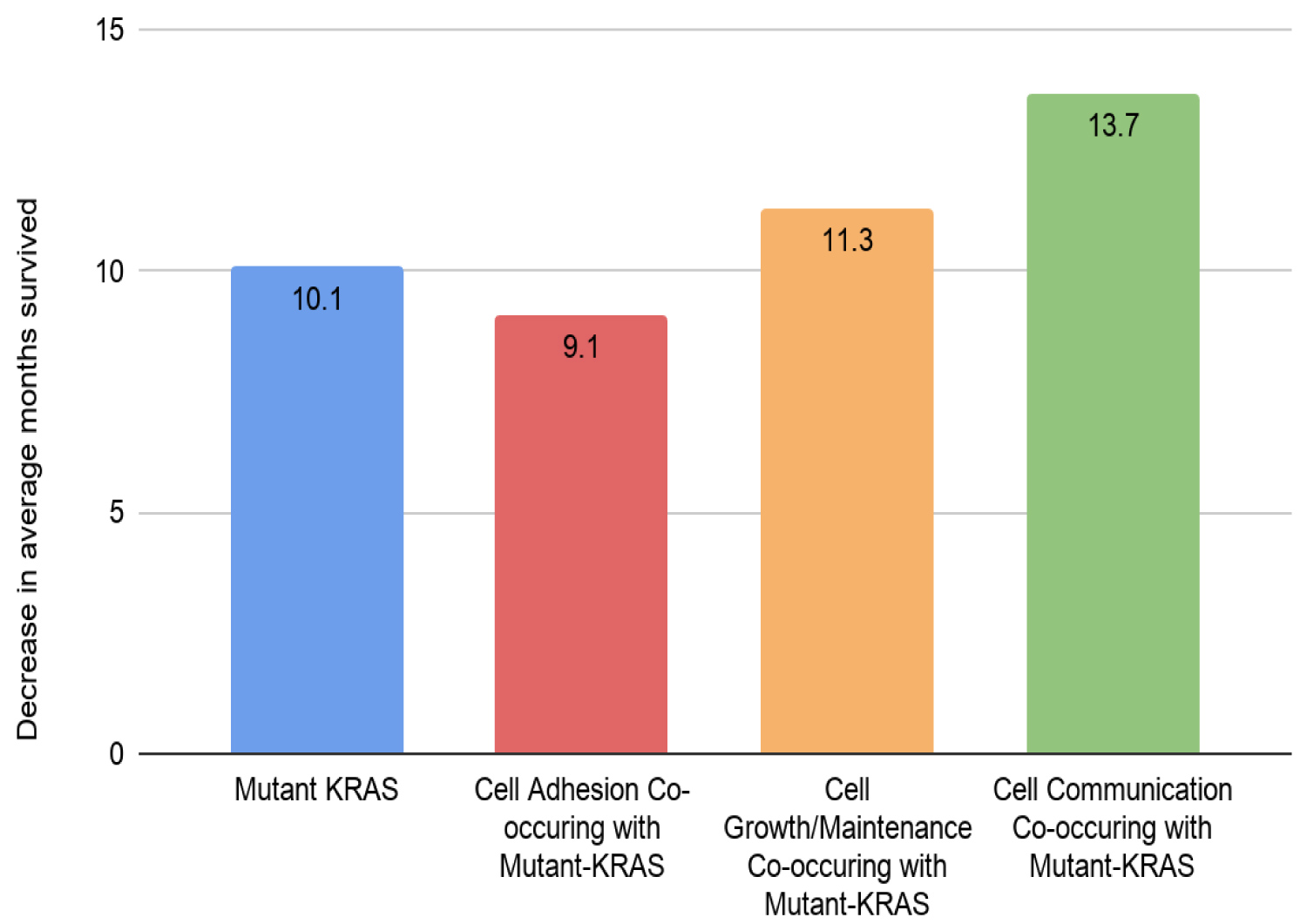
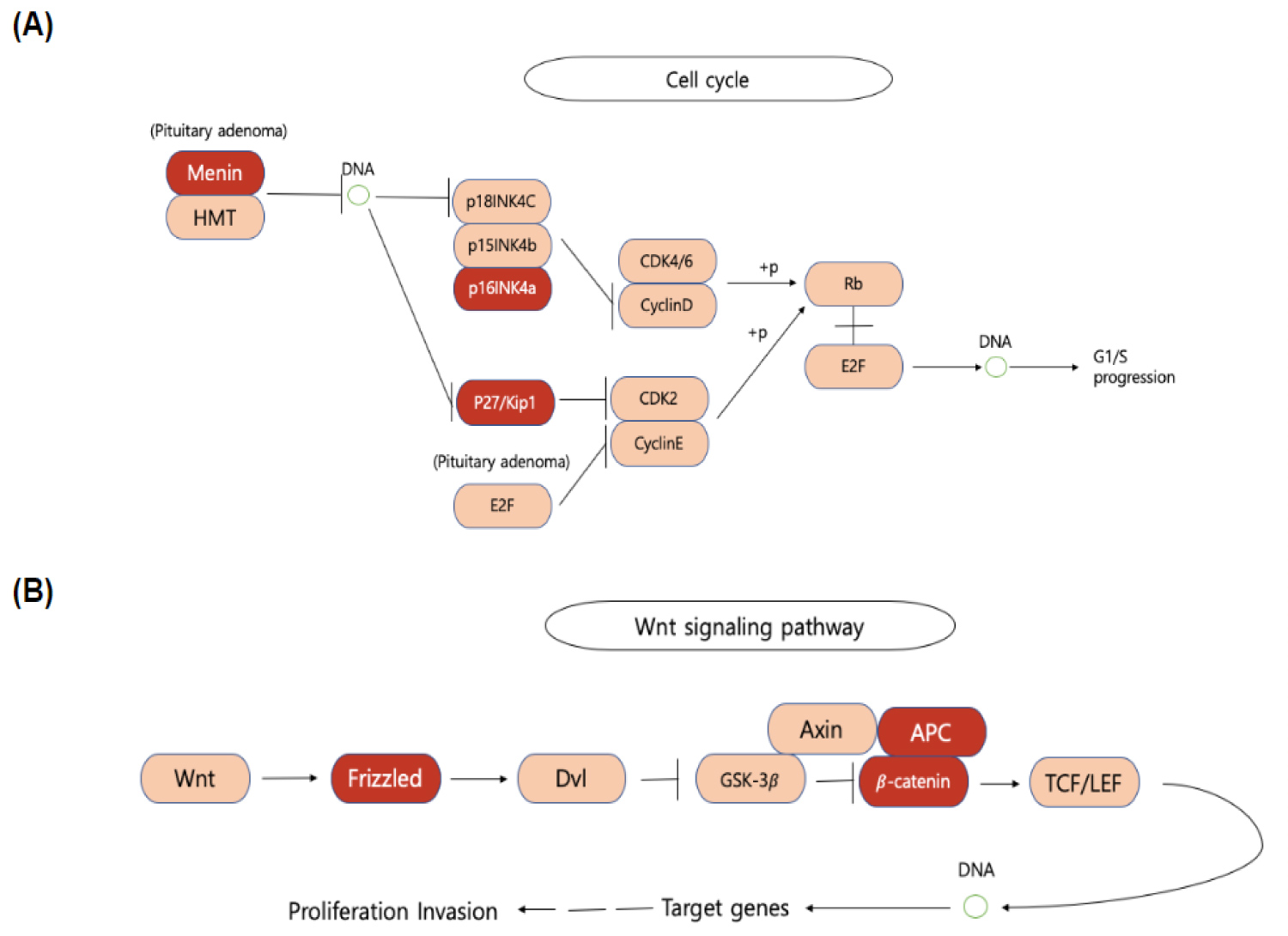
Among the 306 genes found to be co-mutated with mutant-KRAS, CDKN2A, the gene that encodes p16lNK4A, was found to hold a significant role in the progression of the cell cycle (Figure 5). The fact that the mutant form of the CDKN2A gene is found to have the strongest mutation frequency of 16% (Figure 3B) could possibly explain the cell communication group's most significant decrease in the survival time of pancreatic cancer patients in comparison to patients with only the mutant KRAS present (Figure 4). The decrease of patient survival by 13.7 months could be explained by the fact that CDKN2A encodes the protein, p16lNK2A, a protein that holds a significant role within the progression of the cell cycle. Mutations in CDKN2A's vital role as a CDK inhibitor [3] causes a loss of function, leading cells to progress into the S phase without the regulation of CDK levels by the CDKN2A gene. Thus, without the proper regulation of the cell cycle, it could be concluded that tumor formation is highly facilitated by the mutations within the CDKN2A gene.
KEGG mapping analysis also reveals the presence of mutated FZD10, the gene that codes for the frizzled protein, within the Wnt signaling pathway which results in cell proliferation. Frizzled, one of the genes in the cell communication class to be co-mutated with mutant-KRAS, plays a significant role in the reception of the Wnt protein, allowing successive protein chain reactions to occur within the cell. Mutations found in FZD10 could lead to the upregulation of the Frizzled protein, causing the Wnt pathway to occur without the reception of external stimuli. Results from past studies also reveal similar results in the fact that the upregulation of the FZD10 mRNA was found to possibly lead to carcinogenesis in human cells [4]. With the overactivation of the Wnt signaling pathway, cell proliferation could be excessively activated, leading to the development of pancreatic tumors. The following conclusion could perform as an additional explanation of the cell communication group's greatest decrease in pancreatic cancer patient survival time (Figure 4).
Cell growth and maintenance
We researched 7 genes (ASTN1, SYNE1, MYO18B, DNAH7, DYNC2H1, LAMA1) which have mutation tendency more than 2% among 36 genes co-mutated with KRAS. We find out that all genes do not have enough information with cancer or direct relation with pancreatic cancer, but only MYO18B was related to lung cancer.
MY018B is a candidate tumor suppressor gene at chromosome 22q12.1, deleted, mutated, and methylated in human lung cancer. Restoration of MYO18B expression in lung carcinoma suppressed anchorage-independent growth [1]. Therefore, if MYO18B is mutated, it may not play a role as a tumor suppressor anchorage-independent growth of the tumor can be activated. Survival rate of patients with co-occurring gene mutations of cell growth/maintenance and KRAS was higher than the survival rate of those without. However, the data on gene mutation was insufficient in finding the exact relation of mutation and cancer.
Cell adhesion
FunRich analysis revealed the presence of 25 genes of the cell adhesion group to be co-mutated with mutant-KRAS. EMT (epithelial-mesenchymal transition) is known to be a significant process responsible for the invasion of cancer cells. In order to study the impact of co-mutation within the class of adhesion molecules, the adhesion genes were classified into epithelial markers, markers that indicate the presence of cell-binding molecules, and mesenchymal markers, markers that indicate metastatic cancer cells. FunRich analysis found that mutated genes of epithelial markers predominated the list of co-mutant genes. The fact that the majority of mutated cell adhesion molecules were found to be epithelial markers could possibly indicate frequent mutation within the cell-binding molecules, resulting in the cell to cell binds to loosen. Due to such mutations, pancreatic cells could likely express metastatic behavior by spreading to blood vessels that lead to other parts of the body. This could possibly explain the smaller decrease in patient survival time in Figure 4. Co-mutations within the adhesion class may not adversely affect the severity of pancreatic tumors in patients. Rather, the metastatic behavior expressed by the co-mutants could result in the inducing of tumor site development in areas other than the pancreas.
In essence, genes co-occurring with mutant-KRAS display a more adverse impact on patient survival time than simply the presence of a mutant form of KRAS. Cell communication was found to hold the most significant impact on patient survivability, possibly explained by the genes' relation to various pathways. Although previous studies explain the exclusive effects of mutant-KRAS on pancreatic cancer, our study demonstrates that co-occurring genes are also strong candidates in explaining the intricate development of pancreatic cancer.
Method
KRAS co-mutated cancer gene analysis by cBioportal
The cBioPortal is an open-access resource for interactive exploration of multidimensional cancer genomics data sets. 1207 pancreatic cancer patient samples in ten different studies from cBioportal were used to analyze KRAS co-mutated genes. 306 genes have co-occurring mutations in patients with KRAS-mutant pancreatic cancer (p-value < 0.05).
Functional enrichment analysis by Funrich
306 genes were analyzed using the functional enrichment analysis tool (FUNRICH). http://funrich.org/index.html. Gene enrichment analysis was conducted for molecular function and biological process. Pathways were ranked according to the p-value. P < 0.05 was considered statistically significant.
Pathway analysis by KEGG mapper
The KEGG PATHWAY database is deployed as a web application for pathway diagrams. The genes resulting from Funrich were analyzed by the search pathway tool to overlay the co-occurring mutant gene with KRAS-mutation onto biochemical pathways provided by KEGG.
References
- Kent OA (2018) Increased mutant KRAS gene dosage drives pancreatic cancer progression: Evidence for wild-type KRAS as a tumor suppressor? Hepatobiliary Surg Nutr 7: 403-405.
- Soto JL, Cabrera CM, Serrano S, et al. (2005) Mutation analysis of genes that control the G1/S cell cycle in melanoma: TP53, CDKN1A, CDKN2A, and CDKN2B. BMC Cancer 5: 36.
- Terasaki H, Saitoh T, Shiokawa K, et al. (2002) Frizzled-10, up-regulated in primary colorectal cancer, is a positive regulator of the WNT - beta-catenin - TCF signaling pathway. Int J Mol Med 9: 107-112.
- Nishioka M, Kohno T, Tani M, et al. (2002) MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1, deleted, mutated, and methylated in human lung cancer. Proc Natl Acad Sci USA 99: 12269-12274.
Corresponding Author
Oh Dongmyung, Cheong Shim International Academy, South Korea.
Copyright
© 2020 Moon K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.