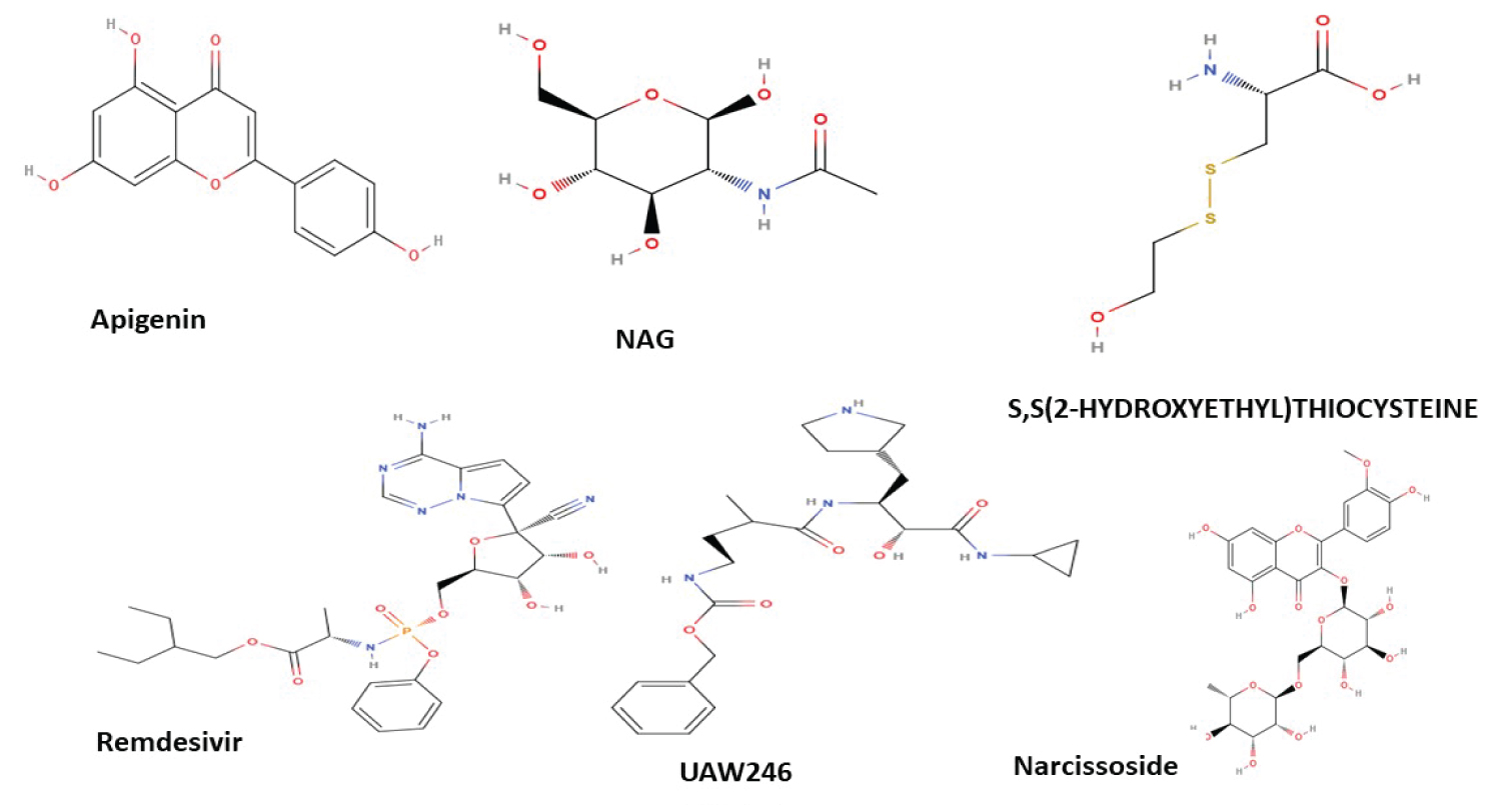
In Silico Identification of Apigenin and Narcissin (Food-Flavonoids) as Potential Targets Against SARS-CoV-2 Viral Proteins: Comparison with the Effect of Remdesivir
Abstract
Background
In this study, we demonstrate the potential role of Narcissin and apigenin, two natural flavonoids found in fruits and foods, as candidate compounds in the treatment against the novel corona virus infection using in silico tools.
Methods
We have used computational molecular docking screening (Molegro Virtual Docker) to study the effect of selected flavonoids on main viral proteins including MERS-COV spike-RBD, RNA-polymerase, viral main protease, and papain-like protease. A grid resolution of 30 A° was used, together with an MM2 force field for energy minimization. Moreover, the MolDock Score and ReRank score were used as scoring functions. The results obtained were compared to those of Remdesivir, a drug under investigation for Covid-19 treatment.
Results
Narcissin, Apigenin and Remdesivir were identified as strong inhibitors of Covid-19 proteins with MolDock scores of: MERS-COV Spike-RBD, PDB: 4KRO (-101.704, -61.069, -15.96), Papain-like protease MERS, PDB: 4P16 (-91.462, -47.314, -43.64), SARS-COV-2 main protease in complex with inhibitor UAW246, PDB: 6XBG (-151.124, -98.20, -150.12) respectively for the three drugs. As far as 7BZ5 (Spike receptor binding domain) and 7BTF (SARS-COV-2-RNA polymerase) proteins are concerned, Remdesivir showed inhibitory effect higher than Narcissin and Apigenin for 7BZ5 (-85.98, -44.78, -60.074), 7BTF (-113.44, -109.32, -93.403) even though the binding affinity towards 7BTF (RNA polymerase) was higher for Narcissin and Apigenin. The results showed that Narcissin has high affinity towards 6XBG as it showed 12 hydrogen bonds with amino acids Phe3, Lys5 and Glu288, while showing 8 hydrogen bonds with 7BTF (RNA polymerase). Apigenin showed 6 hydrogen bonds with amino acids Gly 138 and Glu 288 for 6XBG and 5 hydrogen bonds with amino acids Asn459, Thr462, Arg349 and Met 629 for 7BTF.
Conclusion
From in silico analysis, it was concluded that Narcissin and Apigenin are potential Covid-19 viral inhibitors by directly binding to RNA polymerase and key viral proteins such as the main protease and the spike-RBD. Both flavonoids had high affinity and inhibitory potential than the reference drug used in clinical studies. Narcissin and apigenin are therefore predicted after further confirmation as potent candidate drugs to enter clinical studies as efficient drugs against the novel Covid-19.
Keywords
Apigenin, Narcissin, Remdesivir, Molecular docking, SARS-CoV2 proteins
Introduction
In late 2019, an ongoing outbreak emerged from China Wuhan city. After a noticeably short period of time, what has become a pandemic had spread across the world with an alarming daily increase of cases and deaths [1,2]. As of October 2020, 34,986,502 Covid-19 positive cases were reported with 6 million recovered and 1,034,240 deaths. Fortunately, the World Health Organization (WHO) has taken several measures to help reduce disease transmission and control the outbreak. Nevertheless, the number of cases is increasing, and it appears that well equipped health systems are becoming overwhelmed with the situation. It therefore appears that the identification of safe and effective anti-viral compounds could open the way to the finding of an appropriate treatment by targeting SARS-CoV-2 viral components.
Several viral proteins have been identified and are under investigation in laboratories across the world. Amongst them, the Spike-RBD is the viral protein that binds to the host receptor angiotensin-converting enzyme-2 via its receptor-binding-domain (RBD), and thus is believed to be a major target to block viral entry. The SARS-Cov2 main protease is a key protein in the virus life cycle and is therefore an attractive drug target among coronaviruses since it plays an essential role in processing the polyproteins that are translated from the viral RNA. The RNA polymerase of SARS-CoV-2 plays a pivotal role in viral replication and as such, is a potential target for anti-SARS therapy. Other proteins include virally encoded cysteine proteases.
Natural compounds may provide an alternative approach for the discovery of new drugs with antiviral activities. This is the case of flavonoids which are reported to have antiviral effects against SARS- and MERS-CoV respectively [3]. Coronavirus have always been targets of several active flavonoids by directly inhibiting the protein 3LPro (3-chymotrypsin-like protease) and Papain-like protease (PLPro). This is the case of herbacetin, rhoifolin and pectolonarin, which were found to efficiently block the enzymatic activity of SARS-COV 3CLPro [4]. An in silico and in vitro recent study also reported the inhibitory effect of quercetin, epigallo catechin gallate, Gallocatechin gallate and Rutin on the same protease (3CLPro) [5,6]. Medicinal plants and compounds from Chinese medicine have shown promising effects for SARS-CoV-2 potential treatment. This is the case of Glycyrrhizin which is reported to inhibit the replication of SARS-associated corona viruses in vitro [7,8]. Moreover, high doses of the drug have been used in clinical trials and was reported effective for SARS treatment [9,10]. Herperidin, a flavonoid found in citrus fruits is an inhibitor of ACE2, and thereby blocks the infection of SARS-CoV-2 [11] and cleaves 3CLpro in cell-based assays [12]. Baicalin another flavone showed antiviral activities against 10 clinical isolates of SARS-CoV [13]. Finally, quercetin demonstrated antiviral effect by inhibiting 3CLpro of SARS-CoV [14] and by blocking the entry of SARS-CoV into host cells [15].
For an exceptionally long time, drugs were identified randomly and by chance. Since the development of computed aided methods, the process of drug discovery has been considerably speeded up and Molecular Docking approaches could play a pivotal role in identifying drugs before entering pharmacological and clinical trials.
For centuries, medicinal plants and foods have been reported as major sources of efficient and safe drugs for humans. Amongst them, flavonoids are widely distributed in vegetables, fruits, and leaves. They are known for their anti-inflammatory, antibacterial and anti-tumor effects [16-18]. For instance, Apigenin is a natural flavonoid (4',5,7,-trihydroxyflavone) found in fruits (oranges), vegetables (onions, celery), spices (basil, Oregano) and herbs (tea, beer and red wine). Another flavonoid with potential effect on Covid-19 infection is Narcissin (Isorhamnetin-3-O-rutinoside), also known as Narcissoside, has been identified and isolated from several plants and leaves including the leaves of Manihot escylenta Crantz also known as cassava leaves which are highly consumed in sub-Saharan Africa [19].
So far, no treatment has been found against Covid-19 infection. Remdesivir has entered clinical trials in the United States of America and China, with the purpose of using the drug as a therapeutic for Covid-19. Unfortunately, preliminary clinical trials results suggest only some benefit-risk profile of the drug on severe Covid-19 patients [20]. Indeed, reported adverse effects in patients taking Remdesivir included nausea, acute respiratory failure, and elevated liver enzymes.
In the present work, we have used in silico approach to identify potential viral inhibitors from flavonoids available in daily consumed fruits and vegetables. Cheap, safe, and effective inhibitors of SARS-CoV-2 could help as effective treatment against the Covid-19 pandemic.
Material and Methods
In silico-molecular docking
In this work, we used MVD software which provides very accurate predictions of ligand binding modes (87%) in comparison to other docking softwares [21].
Ligand preparation
The mol files used for docking were obtained from Molview, which was used to draw the structures of ligands when necessary. Narcissin, Apigenin and Remdesivir were identified from PubChem chemical database. The structures were confirmed with Molview software and the energy minimization performed using the MM2 force field and saved as a .MOL format. Energy minimization was done to help the docking programme in the identification of the bioactive conformers. The missing charges and hybridization states of different ligands structure were assigned with the help of the MVD (Molview Virtual Docker) software. The major advantage of MVD is that it can assign the missing bond orders, charges, bonds and hybridization states of the investigated ligands.
Protein preparation
The three-dimensional crystal structure (3D structure) of different receptors was retrieved from the protein data bank (PDB) (http://www.rcsb.org). We have chosen respectively: Papain-like protease of MERS coronavirus (PDB ID: 4P16), MERS-CoV spike-RBD (PDB ID: 4KR0), SARS-CoV-2 main protease (PDB ID: 6XBG), Covid-19 virus spike receptor binding domain (PDB ID: 7BZ5) and SARS-CoV-2-RNA-dependent RNA Polymerase (PDB ID: 7BTF). The proteins had one or two polypeptides and were co-crystallized with ligands. The targets were visually inspected, and reference ligands/inhibitors identified for each receptor. Receptors were prepared for docking by removal of water molecules, ligands, cofactors and assigning bonds, bond order, hybridization and charges using the MVD software [22]. The standard Molegro algorithm was utilized to include the missing charges, protonation states and assigning polar hydrogens to the receptor.
Docking search algorithm and scoring functions
MVD uses PLP (Piecewise Linear Potential) algorithm as scoring function for computational screening. In this study, the MolDock simplex evolution search algorithm was used for docking. In practice, the number of receptor cavities was limited to four and using the cavity prediction wizard, the cavity with largest volume was selected for simulations. Docking of compounds in different receptors was performed and the best poses generated were used based on the MolDock and ReRank scores.
Parameters for scoring functions
MVD is based on differential evolution algorithm called Moldock. In practice, MVD software uses two scoring functions: The MolDock score and the ReRank score, where the MolDock score is an E score (docking scoring function) defined as: E score = E inter + E intra.
E inter: Sum of ligand-protein interaction energy, ligand-water interaction energy and ligand-cofactor interaction energy [23].
E intra: Internal energy of the ligand.
The Eplp term means piecewise linear potential. It uses two different parameters, one for the estimate of van der walls interactions between atoms and another for the potential of hydrogen bons, describing the electrostatic interactions between charged atoms. Eintra is calculated as follows:
The first term in the above equation calculates all energies involving pairs of atoms of the ligand, except those associated with two bonds. The second term is the torsional energy. The last term, E clash, assigns a penalty of 1000 Kcal/mol if the distance between two heavy atoms is smaller than 20 A [21].
ReRank score provides an estimation of the ligand-receptor interaction strength [24].
Finally, a GRID function of 0.30 A° and a binding site radius of 12 A° were used with respect to the origin of different cavities. Ten runs were used for the searching algorithm with a maximum of 1500 iterations with a total population size of 50 applied. The energy threshold for the minimized final orientation was 100 [25]. The binding free energy, described as a sum of the intermolecular interactions between the ligand and the receptor and the steric energy of the ligand was given in Kcal/mol.
Results
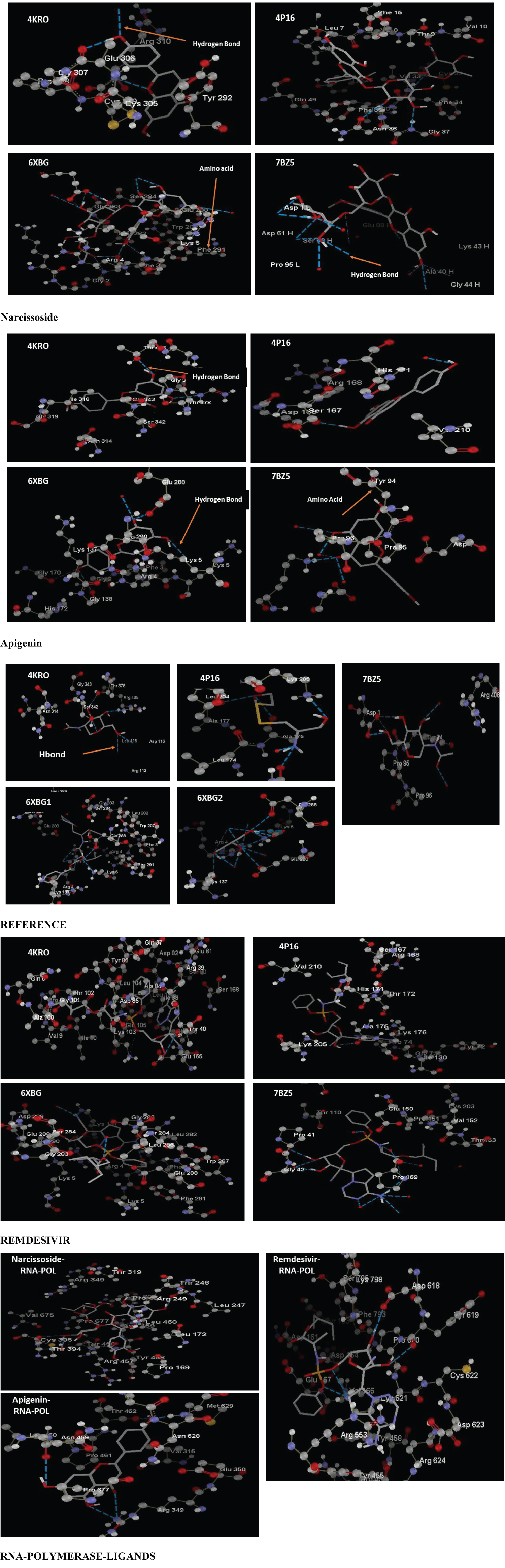
A considerable number of flavonoids including apigenin, baicalin, cis resveratrol, epicatechin, epigallocatechin gallate, hesperidin, icariin, oridonin, quercetin, and rutin were identified and docked using the MolDock virtual screening software (data not shown). The Docking scores obtained showed an extremely low binding of the majority of flavonoids tested. However, interesting scores were obtained with Apigenin and Narcissin which were further considered for docking analysis in this study. Identified flavonoids as well as receptor's ligands/inhibitors 2D structures are represented in Figure 1. The results obtained are represented in Table 1, Table 2, Table 3 and Table 4, indicating that Narcissin, Apigenin and Remdesivir perfectly bind at the active sites of investigated receptors. As far as 4KRO is concerned, the highest binding score was obtained with Narcissin, followed by Apigenin and Remdesivir (Table 2 legend). The binding involves interactions with 15 amino acids which are listed in Table 3. The highest binding score was the one obtained with Narcissin for the receptors 4P16 and 6XBG. For 4P16, ten amino acids were involved in the binding site (listed in Table 2). For 6XBG, the binding site involved fourteen amino acids (Table 3). Remdesivir showed the highest binding scores for 7BTF and 7BZ5 with seventeen and eleven amino acids involved in the binding mechanism respectively (Table 3).
UAW246 is the reference inhibitor for 6XBG. Remdesivir and Narcissoside showed the highest binding energies in comparison with the reference inhibitor of the enzyme, as shown in Table 2. The score obtained with Apigenin was not far from the one obtained with UAW246. However, five amino acids were changed between the inhibitor and Remdesivir. It was observed that UAW246 interacted with Glu290, Gly138, Gly283, Lys137 while Remdesivir interacted with Gly2, Gly283, Ile281, Leu286, Trp207. As far Apigenin is concerned, UAW246 interacted with Gly283, Phe291 and Ser284, while Apigenin interacted with Gly2, Lys5, Gly170, His172, Ser139 and Val171.
For 6XBG, the docking scores obtained with Moldock virtual docker showed that Remdesivir had a MolDock score of -150.12, Rerank score of -117.1 and Hbond -8.774, while Apigenin had a MolDock score of -98.20, Rerank score of -82.98 and Hbond -3.62 and Narcissin a MolDock score of -151.12, Rerank score of -116.1 and Hbond -11. With the reference inhibitor, the results showed a MolDock score of -114.93, Rerank score of -98.25 and Hbond -11.062. With 7BTF docked with Narcissin, the MolDock score was -109.321, Rerank score of -41.1 and Hbond -7.8. The same enzyme docked with Remdesivir showed a MolDock score of -113.44, ReRank score -95.23 and Hbond -6.100. Therefore, Narcissin and Apigenin showed potent inhibitory effect on 6XBG, 4P16 and 4KR0 which is greater than that of Remdesivir. Both flavonoids almost showed similar inhibitory effect on 7BTF in comparison to the result obtained with Remdesivir (Figure 2).
Discussion
The pandemic caused by SARS-CoV-2 remains a major health issue due to its devastating effects on health and economic systems worldwide. By end January 2020, the World Health Organization declared Covid-19 outbreak as a public health emergency [26]. Indeed, within a few months, the disease which originated from Chinese Wuhan Province had spread across the globe, reaching the level of a pandemic [27]. The clinical symptoms experienced by Covid-19 patients include fatigue, fever, cough, and the disease could progress to more serious outcomes including the acute respiratory syndrome [28]. Until now, no treatment has been found despite multiple drugs and potential vaccines under active investigation. Our present work is aimed to find out potent inhibitors of SARS-CoV-2 viral proteins from daily consumable fruits and vegetables.
In the first part of our investigation, several flavonoids were tested in in silico tests to identify potential inhibitors of SARS-CoV-2 proteins. Amongst them, Apigenin and Narcissin showed interesting binding scores and were selected for further investigations. The results obtained were analyzed and compared to those of Remdesivir, an Anti-Covid drug which has entered clinical studies in China and the USA (United States of America) and reported to shorten the time to recovery of Covid-19 patients [29]. Apigenin is a natural flavone widely found in plants such as celery, parsley and chamomile [30]. Apigenin has gained interest in recent years due to its health-promoting effects on inflammation, cancer, and inflammation. Some flavonoids like kaempferol have shown potent inhibitory effect on SARS-CoV-2 in docking studies [31]. Other anti-nCov drugs have been identified from Indian spices as targeting SARS-CoV-2 main protease [32]. In a recent study, green tea polyphenol constituents (epigallocatechin gallate, epicatechin gallate and gallocatechin-3-gallate) showed inhibitory effects on SARS-CoV-2 main protease in an in silico docking and molecular dynamics simulation study [33]. Inhibitory effects of Narcissin were reported against 6W63, the main protease inhibitor [22]. Narcissoside is found in cassava leaves, a meal highly appreciated in Cameroon and other sub-Saharan countries.
Protein-ligand interaction studies are necessary to understand the mechanism of action of potential drugs as they provide the basis for the design and discovery of drug targets. In this study, different ligands were docked with 6XBG receptor, which is an attractive drug target for lead compounds against the virus. Moreover, the results obtained were compared to those of UWA246, the standard inhibitor of the enzyme. The three ligands had bound the active cavity of the receptor and exhibited significant binding with amino acids present in the receptor's pocket. The binding energy of Narcissin with 6XBG with amino acids Arg4 (-15.267), Glu288 (-11.448), Glu290 (-0.787), Gly2 (-2.734), Leu282 (-18.0015), Lys5 (-9.549), Phe3 (-21.644) were compared with the binding energy of Apigenin with amino acids Arg4(-27.189), Gly2(-4.039), Leu282(-1.102), Phe3(-9.587), Glu288(-5.574), Glu290(-6.248), Lys5(-7.869), and Remdesivir Arg4(-18.026), Glu288(-12.053), Leu282(-17.503), Lys5(-1.835,), Phe3(-10.641), Phe291(-4.840), Ser284(-5.863). With the reference inhibitor, amino acids included: Arg4(-6.587,), Glu288(-11.929), Glu290(-1.468), Leu282(-8.611), Lys5(-18.6314), Phe3(-6.096), Phe291(-4.667). The results obtained show that Narcissin, Remdesivir and Apigenin require less energy to bind to 6XBG receptor than the known inhibitor of the receptor. Analysis of interactions between the receptor and different ligands showed higher affinity with Narcissin with twelve hydrogen bonds with Phe3, Lys5, Glu288 while showing three hydrogen bonds with the inhibitor UAW246 Arg4(B), Lys5(A), Lys137(A). Indeed, hydrogen bonds are important contributors to the stability and the specificity of receptor-ligand interactions. The protein showed 6 hydrogen bonds with Apigenin Gly138(B), Glu288(B) and eight hydrogen bonds with Remdesivir 8 Phe3, Arg4, Glu288.Narcissoside and Apigenin showed higher affinity than Remdesivir for 7BTF (RNA polymerase) as they established for Narcissin 8 hydrogen bonds (Arg249, Thr246, Pro677, Tyr456, Thr394), Apigenin 5 hydrogen bond (Thr462, Met629, Asn459, Arg349) and 4 hydrogen bonds for Remdesivir (Tyr619, Asp618, Lys621). The elevated presence of Narcissin and Apigenin in consumed foods may explain why the pandemic seems less devastating in certain regions of the world like in Sub-Saharan Africa.
Preclinical evidence has shown strong antioxidant, anti-inflammatory and anti-neoplastic effects, even though no clinical trial have directly tested the effect of apigenin in humans. The drug is generally safe but excessive amounts may cause drug interactions due to CYP2C9, an enzyme involved in drug metabolism but may interfere with other drugs metabolism. Moreover, no toxicity has been reported in humans and in rodents even at high doses [34]. Apigenin is classified in class II per the Biopharmaceutics classification system and is described as a drug with poor solubility but high intestinal permeability (ref). However, different strategies could be used to solve the solubility issue. For instance, novel carriers would need to be developed in order to enhance its oral bioavailability [30].
Narcissoside has anti-oxidant and antiproliferative properties against cancer [35]. To the best of our knowledge, toxicological and bioavailability properties of Narcissoside have not been fully investigated. However, due to their pleiotropic activities, the lack of systemic toxicity and the prominent results obtained in silico on the inhibition of SARS-CoV-2 viral components, apigenin and Narcissoside may represent lead compounds to be investigated in future clinical trials for Covid-19 treatment. The compounds will therefore need to be formulated in suitable dosage forms to ensure a maximal bioavailability.
Conclusion
The severity of the new Covid-19 infection urges the discovery and finding of drugs to enter clinical studies. Finding lead compounds from available foods and fruits may help identify safe, cheap, and effective treatment against the virus. In the present research work, Narcissin and Apigenin were studies in silico for their binding effect on main SARS-CoV-2 viral proteins.The results obtained show that Narcissin and Apigenin have higher binding affinity than Remdesivir at fit perfectly at the active sites of receptors 6XBG (main viral protease) and 7BTF (RNA Polymerase). For 7BTF, Narcissin showed 8 hydrogen bonds, Apigenin 5 hydrogen bonds and Remdesivir 4. Therefore, further investigations are necessary to evaluate the effectiveness and potential of Narcissin and Apigenin in clinical trials against the new SARS-CoV-2 virus.
Conflict of Interest
The authors of this research declare that they have no conflict of interest.
References
- Wu F, Zhao S, Yu B, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265-269.
- Baloch S, Baloch MA, Zheng T, et al. (2020) The coronavirus disease 2019 (COVID-19) pandemic. Tohoku J Exp Med 250: 271-278.
- Solnier J, Fladerer JP (2020) Flavonoids: A complementary approach to conventional therapy of COVID-19? Phytochem Rev 18: 1-23.
- Jo S, Kim S, Shin DH, et al. (2020) Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem 35: 145-151.
- Nguyen TTH, Woo HJ, Kang HK, et al. (2012) Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol Lett 34: 831-838.
- Cherrak SA, Merzouk H, Mokhtari SN (2020) Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies. PLoS One 15: e0240653.
- Cinatl J, Morgenstern B, Bauer G, et al. (2003) Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361: 2045-2046.
- Hoever G, Baltina L, Michaelis M, et al. (2005) Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J Med Chem 4: 1256-1259.
- Lu H, Huo N, Wang G, et al. (2003) Clinical observation of therapeutic effect of compound glycyrrhizin on SARS. China Pharmacy.
- Ma W, Chen GF, Li TS, et al. (2003) Analysis of chest X-ray manifestations in 118 patients with severe acute respiratory syndrome. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue 15: 338-342.
- Chen H, Du Q (2020) Potential natural compounds for preventing SARS-CoV-2 (2019-nCoV) infection. Medicine & Pharmacology.
- Lin CW, Tsai FJ, Tsai CH, et al. (2005) Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res 68: 36-42.
- Chen F, Chan KH, Jiang Y, et al. (2004) In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol 31: 69-75.
- Chen L, Li J, Luo C, et al. (2006) Binding interaction of quercetin-3-ß-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: Structure-activity relationship studies reveal salient pharmacophore features. Bioorg Med Chem 14: 8295-8306.
- Wu CY, Jan JT, Ma SH, et al. (2004) Small molecules targeting severe acute respiratory syndrome human coronavirus. PNAS 101: 10012-10017.
- Pietta PG (2000) Flavonoids as antioxidants. J Nat Prod 63: 1035-1042.
- Tsou LK, Lara TM, Rosefigura J, et al. (2016) Antibacterial flavonoids from medicinal plants covalently inactivate type III protein secretion substrates. J Am Chem Soc 138: 2209-2218.
- Seelinger G, Merfort I, Wölfle U, et al. (2008) Anti-carcinogenic effects of the flavonoid luteolin. Molecules 13: 2628-2651.
- Tao H, Cui B, Zhang H, et al. (2019) Identification and characterization of flavonoids compounds in cassava leaves (Manihot esculenta Crantz) by HPLC/FTICR-MS. International Journal of Food Properties 22: 1134-1145.
- Davies M, Osborne V, Lane S, et al. (2020) Remdesivir in treatment of COVID-19: A systematic benefit–risk assessment. Drug Saf 43: 645-656.
- Thomsen R, Christensen MH (2006) MolDock: A new technique for high-accuracy molecular docking. J Med Chem 49: 3315-3321.
- Dubey K, Dubey R (2020) Computation screening of narcissoside a glycosyloxyflavone for potential novel coronavirus 2019 (COVID-19) inhibitor. Biomedical Journal 43: 363-367.
- Heble NK, Mavillapalli RC, Selvaraj R, et al. (2016) Molecular docking studies of phytoconstituents identified in Crocus sativus, Curcuma longa, Cassia occidentalis and Moringa oleifera on thymidylate synthase - An enzyme target for anti-cancer activity. J App Pharm Sci 6: 131-135.
- Madhuri M, Prasad C, Rao AV (2014) In silico protein-ligand docking studies on thiazolidinediones as potential anticancer agents. International Journal of Computer Applications 95: 13-16.
- Kaushik P, Lal KS, Rana AC, et al. (2014) Pharmacophore modeling and molecular docking studies on Pinus roxburghii as a target for diabetes mellitus. Advances in Bioinformatics.
- Pfefferbaum B, North CS (2020) Mental health and the Covid-19 pandemic. N Engl J Med 383: 510-512.
- Yang B, Zhu Y (2020) Epidemiology and treatment of novel coronavirus pneumonia (COVID-19) in children and adults.
- Guo YR, Cao QD, Hong ZS, et al. (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- an update on the status. Mil Med Res 7: 11.
- Beigel JH, Tomashek KM, Dodd LE, et al. (2020) Remdesivir for the treatment of Covid-19 - preliminary report. N Engl J Med 383: 992-994.
- Tang D, Chen K, Huang L, et al. (2017) Pharmacokinetic properties and drug interactions of apigenin, a natural flavone. Expert Opin Drug Metab Toxicol 13: 323-330.
- Hamza M, Ali A, Khan S, et al. (2020) nCOV-19 peptides mass fingerprinting identification, binding, and blocking of inhibitors flavonoids and anthraquinone of Moringa oleifera and hydroxychloroquine. Journal of biomolecular structure & dynamics. J Biomol Struct Dyn 2020: 1-11.
- Umesh U, Kundu D, Selvaraj C, et al. (2020) Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J Biomol Struct Dyn, 1-9.
- Ghosh R, Chakraborty A, Biswas A, et al. (2020) Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn, 1-13.
- Venigalla M, Sonego S, Gyengesi E, et al. (2016) Novel promising therapeutics against chronic neuroinflammation and neurodegeneration in Alzheimer's disease. Neurochem Int 95: 63-74.
- Calzada F, Valdés M, Merlin VI, et al. (2020) Phytochemical analysis, antipropulsive and antilymphoma activities of leaves extract of Annona cherimola miller. Pharmacognosy Magazine 16: 317-320.
- Seadawy MG, Gad AF, Elhoseny MF, et al. (2020) In vitro: Natural compounds (thymol, carvacrol, hesperidine, and thymoquinone) against SARS-COV2 strain isolated from egyptian patients. bioRxiv.
- Kim DW, Seo KH, Curtis LMJ, et al. (2014) Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzyme Inhib Med Chem 29: 59-63.
- Russo M, Moccia S, Spagnuolo C, et al. (2020) Roles of flavonoids against coronavirus infection. Chemico-Biological Interactions 328: 109211.
Corresponding Author
Dr. Vincent Brice Owona Ayissi, Laboratory of Molecular Pharmacology and Toxicology, Department of Biochemistry, University of Yaounde 1, Yaounde, Cameroon.
Copyright
© 2021 Owona VBA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.