Bee Venom Melittin Modulates Phospholipase A2 Activity by Affecting Substrate Interface on the Surface of Phosphatidylcholine Membrane
Abstract
Melittin is a small amphipathic cationic membrane-active protein in bee venom. Apart from being a key agent in envenomation, melitt in is a physiologically active protein with important pharmacological properties that include anti-cancer and antimicrobial activities. Perhaps melittin's most important property is its ability to modulate the activity of endogenous phospholipase A2 (PLA2) both in vivo and in vitro. Understanding the underlying mechanism in PLA2 modulation is crucial to developing novel pharmaceuticals to treat diseases associated with abnormal increases or decreases in PLA2 activity. Snake venom PLA2s (svPLA2s) have high structural homology and functional activities to endogenous PLA2s and are commonly used to study PLA2 modulation. In this study we employed differential scanning microcalorimetry and continuous fluorescence displacement assays to investigate the changes induced by melittin on the packing of phosphatidylcholine (PC) in liposome membranes and how these changes affect the esterase activity of acidic and basic PLA2 enzymes from rattlesnake venom. By employing electron paramagnetic resonance spectroscopy of spin labeled melittin we examined the association of melittin with svPLA2s in buffers of various pH. We also used AutoDock computation to simulate the interaction of melittin with PC polar headgroups in order to identify hypothetical amino acid residues on the molecular surface of melittin that interact with charged and polar headgroups of PC. Overall, the analysis of data obtained in this study and the data previously published by us and others allowed us to present molecular details on how melittin disturbs PC packing in a bilayer of liposomes to make the phospholipid substrate interface on the membranes surface more conducive to the esterase activity of PLA2.
Keywords
Bee venom melittin, Rattlesnake venom phospholipase A2, Phosphatidylcholine liposomes, Differential scanning microcalorimetry, Continuous fluorescence displacement, Electron paramagnetic resonance, AutoDock computation
Abbreviations
DSMC: Differential scanning microcalorimetry; EPR: Electron paramagnetic resonance; NMR: Nuclear magnetic resonance; HPLC: High performance liquid chromatography; SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresis; PLA2: Phospholipase A2; DAUDA: 11-(dansylamino) undecanoic acid; DMPC: Dimyristoylphosphatidylcholine; DOPC: Dioleoyl phosphatidylcholine; PC: Phosphatidylcholine; FABP: Fatty acid binding protein; PITP: Phosphatidylinositol transfer protein; Cmm: Crotalus molossus molossus
Introduction
Bee venom melittin is an amphipathic cationic membrane-active polypeptide of 26 amino acid residues [1]. In conjunction with bee venom phospholipase A2 (PLA2) melittin exhibits an array of pharmacological activities including anti-inflammatory [2], anti-arthritic [3], anti-nociceptive [3], antimicrobial [4], and anticancer [5] effects. When melittin binds to membrane surfaces it induces changes in the membrane structural integrity including formation of pores, formation of vesicles and fusion of membranes [6,7]. These changes in membrane structure promote changes in membrane potential [8], in aggregation of membrane proteins [9], and in induction of hormone secretion [10].
Melittin-induced alteration of membrane structure stimulates various enzymes such as adenylate cyclase [11], phospholipase D [12], phospholipase C [13], G-protein [14] and protein kinase C [15]. The molecular mechanisms underlying the diverse effects of melittin are not fully understood.
The activity of PLA2 is modulated by melittin, a process important for its pharmacological implications, and it is tested in vitro and in vivo [16]. PLA2, an enzyme that catalyzes the hydrolysis of the sn-2 position of glycerophospholipids to yield lysophospholipids and fatty acids, is present in intracellular and extracellular forms in virtually every tissue and cell type [17]. The produced fatty acids serve as substrates in the synthesis of eicosanoids, which are acute mediators of inflammation and other pathophysiological processes [18].
An understanding of the regulatory mechanisms of sPLA2 is important for treating pathologies resulting from inflammatory diseases like arthritis, and neurological diseases, cardiovascular diseases and cancer [19]. Understanding the mode of action of endogenous PLA2modulators is of a great importance for the engineering of synthetic modulators, especially for developing efficient therapeutic agents.
Snake venom PLA2s exhibit a high degree of structural homology with mammalian endogenous PLA2s. Both snake venom and mammalian PLA2s potentiate very similar pathophysiological and pharmacological reactions [20]. The physiological/pharmacological reactions and esterase activities of both types of PLA2s are modulated by melittin both in vitro and in vivo [5,21,22]. Despite the dedicated efforts devoted to studies involving melittin and PLA2s over the course of several decades, a molecular mechanism for melittin modulation of the physiological and esterase activities of both mammalian and snake venom PLA2 is not totally understood.
The outer leaflet of plasma cell membranes is made of phosphatidylcholine (PC) [23]. Although the polar head of PC including basic choline and acidic phosphate moieties is electrically neutral, the choline moiety of PC is exposed on the outer surface of plasma membrane to repel basic proteins outside cells away from the cell surface [23]. Melittin is the only known basic protein toxin that can penetrate the positively charged field on the cell membrane surface [24], but the amino acid residues on molecular surface of melittin that bind to the cell membrane surface are not yet identified. In this study we report for the first time a set of amino acid residues on melittin surface that are putatively involved in binding to the PC membrane surface. We also report the effects of melittin on the packing of PC in model membranes and on the esterase activity of acidic and basic PLA2 enzymes isolated from Cmm (Northern black-tailed rattlesnake) venom. Our results in this study show that melittin modulates PLA2 esterase activity, not through the direct interaction with the enzyme, but through changes induced by melittin on the interface of the PC substrate to increase activity of acidic PLA2 and to decrease activity of basic PLA2. These findings may prove important in the development of melittin based pharmaceutical agents.
Materials and Methods
Reagents
Melittin, dimyristoylphosphatidylcholine (DMPC), 4-(2-iodoacetamido)-TEMPO, oleic acid and venom from Cmm were purchased from Sigma Chemical Co. (St. Louis, MO). 11-(dansylamino) undecanoic acid (DAUDA) was from Molecular Probes (Junction City, OR). Rat liver fatty-acid binding protein (FABP) was a gift from Dr. S. E. Mansurova of Lomonosov Moscow State University. Acidic and basic PLA2 enzymes were isolated from Cmm rattlesnake venom as previously described [25]. Melittin was purified from the trace PLA2 contamination by cation exchange HPLC on a SCX 83-C-13-ET1 Hydropore column (Rainin Instrument,Woburn, MA) as previously described [26]. The identity and purity of acidic and basic PLA2 enzymes and purity of melittin were determined by reducing SDS-PAGE and by isoelectric focusing. All other reagents used in this study were purchased from Sigma Chemical Co. (St. Louis, MO, USA).
Differential scanning microcalorimeter assay
Large unilamellar liposomes at a DMPC concentration of 3 mM, used for differential scanning microcalorimetry (DSMC), were prepared at 30 ℃ by using the ether evaporation method as previously described [27] in a buffer solution containing 10 mM Tris-HCl, pH 7.0, 0.1 M NaCl and 0.1 mM CaCl2. Liposome samples in one group were treated with either 3 × 10-5 M melittin for 1 min or 10-8 M acidic or basic PLA2 for 5 min at 37 ℃. 5 mM EDTA was then added to the liposomes prior to recording the calorimetric curves. Liposome samples in the second group were pretreated at 37 ℃ with 3 × 10-5 M melittin for 1 min and post-treated with either 10-8 M acidic or basic PLA2 for 5 min, then 5 mM EDTA was added prior to the recording of calorimetric curves. Liposome samples in the third group were pretreated for 1 min with either 10-8 M acidic or basic PLA2 at 37 ℃ and post-treated with 3 × 10-5 M melittin for 5 min. 5 mM EDTA was then added prior to the recording of the calorimetric curves. Control liposome samples in aqueous solution containing 3 mM DMPC, 10 mM Tris-HCl, pH 7.0, 0.1M NaCl, 0.1 mM CaCl2 and 5 mM EDTA were incubated at 37 ℃ for 6 min in the absence of melittin and PLA2. Calorimetric curves of liposome samples were monitored at a recording rate of 1 ℃ per min using a differential scanning microcalorimeter DASM-4 (Saint-Petersburg, Russia) equipped with the software to calculate enthalpy change (∆H) and the main phase transition peak width at its half-height (). The instrumental base line calibration mark was obtained by scanning at 50 mW and DT = 4 as previously described [28]. Each liposome sample for the DSMC assay was prepared and tested in triplicate. Each data point for ∆H and values is a mean of separate experiments with the SD ± 1.4% of the means.
PLA2 assay
PLA2 activity was measured by a continuous fluorescence displacement assay [29]. Large unilamellar liposomes at a DMPC concentration of 3 mM were prepared at 30 ℃ by the ether evaporation method [27] and were used as substrates for PLA2. Aqueous solutions of liposomes consisted of 10 mM Tris-HCl (pH 7.0), 0.1M NaCl and 0.1 mM CaCl2. Acidic or basic PLA2 enzymes (10-8 M) were incubated with liposome samples for 5 min at 37 ℃. In some assays, liposomes were pre- or post-treated with 3 × 10-5 M melittin the same way as described for the DSMC assay above. PC hydrolysis was terminated by addition of 5 mM EDTA.
Control samples were incubated in the absence of melittin or PLA2. The liposomes (1 ml) were dissolved by vortex shaking in 5 mM Triton X-100 and then mixed with 1 ml of 10 mM Tris-HCl buffer (pH 7.0) containing 25 μg of rat liver FABP. The mixture was added to a thermostatically regulated (37 ℃) fluorimeter cell containing 30 μl of 0.1 M DAUDA in methanol. The solution was excited by pulsed laser at 350 nm, and the fluorescence of DAUDA was measured at 500 nm by using a Hitachi F200 fluorimeter. The excited state lifetime of DAUDA, that reflected an equilibrium between DAUDA and free fatty acids binding to rat liver FABP, was estimated from the time dependence of the attenuation of the probe glow using semilogarithmic coordinates. A standard curve of the lifetime of the excited state of DAUDA as a function of free fatty acid concentration was prepared using defined concentrations of oleic acid. PLA2 activity was expressed as μmoles fatty acid released per 10-11 M moles of enzyme. Each sample was prepared and measured at least in triplicate. The SD was always within ± 2.3% of the means.
EPR of spin labeled melittin study
Melittin was covalently labeled with 4-(2-iodoacetamido)TEMPO according to the Sigma Chemical (St. Louis, MO) protocol with minor modifications as previously described [26,30]. Labeling melittin at a threefold molar excess of spin label over melittin, reduced melittin cytotoxicity by 7% on human lymphocytes (assessed by trypan blue exclusion). The melittin concentration was 10-5 M. Unreacted spin label was removed from the reaction mixture by Sephadex G-100 gel filtration.
From the amount of spin label removed from the reaction mixture we estimated that one mole of spin label covalently binds to one mole of melittin at a threefold molar excess of spin label over melittin. A highly resolved EPR signal was recorded from 0.1 ml of buffer with 10-5 M spin-labeled melittin in EPR tube. Spin-labeled melittin (10-5 M) was incubated for 5 min at 37 ℃ in the presence of 4 × 10-5 M of either acidic or basic PLA2 in buffers (2 mM Tris-HCL, 0.1M NaCl) of various pHs. Spin-labeled melittin in the same buffers without PLA2 served as controls. The EPR spectra of spin-labeled melittin were recorded with a Varian E-4 spectrometer at modulation amplitudes not exceeding 2 × 10-4 T and resonator input power not exceeding 20 mW. Each sample was prepared and recorded at least in triplicates. The differences in hyperfine splitting and intensity of resonance peaks of EPR spectra from different preparations for each sample were always within 0.5%.
Molecular docking
Docking of PC with the molecular surface of melittin was done by using the AutoDockVina Version 4.2 program and using PDB coordinates of PC (structure of PITP complexed to DOPC - PDB ID# 1T27) and melittin (PDB ID# 2MLT) by applying a similar methodology and parameters as previously published [31], but containing the following minor modifications required for this study. The PC virtual molecule was further edited to remove the alkyl chains using Avogadro as previously published [32], and the overall chargeswere checked and energy-minimized using AutoDockVina. A grid box was set up with the following dimensions: Center of x = 28.91; center of y = -2.077; center of z = 17.956; length of x = 100 Å; length of y = 72 Å; and length of z = 48 Å. The setting for exhaustiveness was set up as 16, which gave us consistent results in at least three sets of docking for PC polar head and melittin pair. Following each AutoDock run, the best nine docked conformations were analyzed for ionic, ion-polar and hydrogen bond interactions between the PC polar head groups and charged and polar amino acid groups of melittin by using Python Molecular Viewer (MGL Tools, The Scripps Research Institute).
Statistics
The data points in this study are expressed as means ± SD from at least three independent experiments. Data were analyzed by Student's t test (two-tailed) for single comparisons. Multiple group comparisons were done by performing one-way ANOVA followed by Bonferroni-corrected Tukey's test. P values less than 0.05 were considered statistically significant.
Results
Melittin is a protein with a straight-line 3D structure of alpha-helical rod that slightly bends at amino acid residue P14 [33]. It has been shown previously that melittin binds to the PC membrane surface with the helical axis parallel to the bilayer plane [33]. Melittin stays in the interface between lipid polar head region and non-polar area of alkyl chains and does not penetrate into the inner monolayer [33,34] which increases the surface area of outer monolayer over the inner monolayer causing an asymmetrical interfacial area tension. At a lipid to melittin molar ratio exceeding 100 to 1, the build-up in interfacial tension is released through the formation of stable pores in which melittin helices start changing initial parallel orientation to perpendicular orientation with respect to the membrane plane [34]. In this study we have used a lipid to melittin molar ratio 100 to 1 as we wanted to investigate the effects of melittin on the PC membrane packing order and on PLA2 esterase activity at a threshold at which a disturbed PC bilayer packing starts changing to a nonlamellar PC formation of stable pores.
Effects of melittin on DMPC membrane packing and esterase activity of acidic and basic PLA2
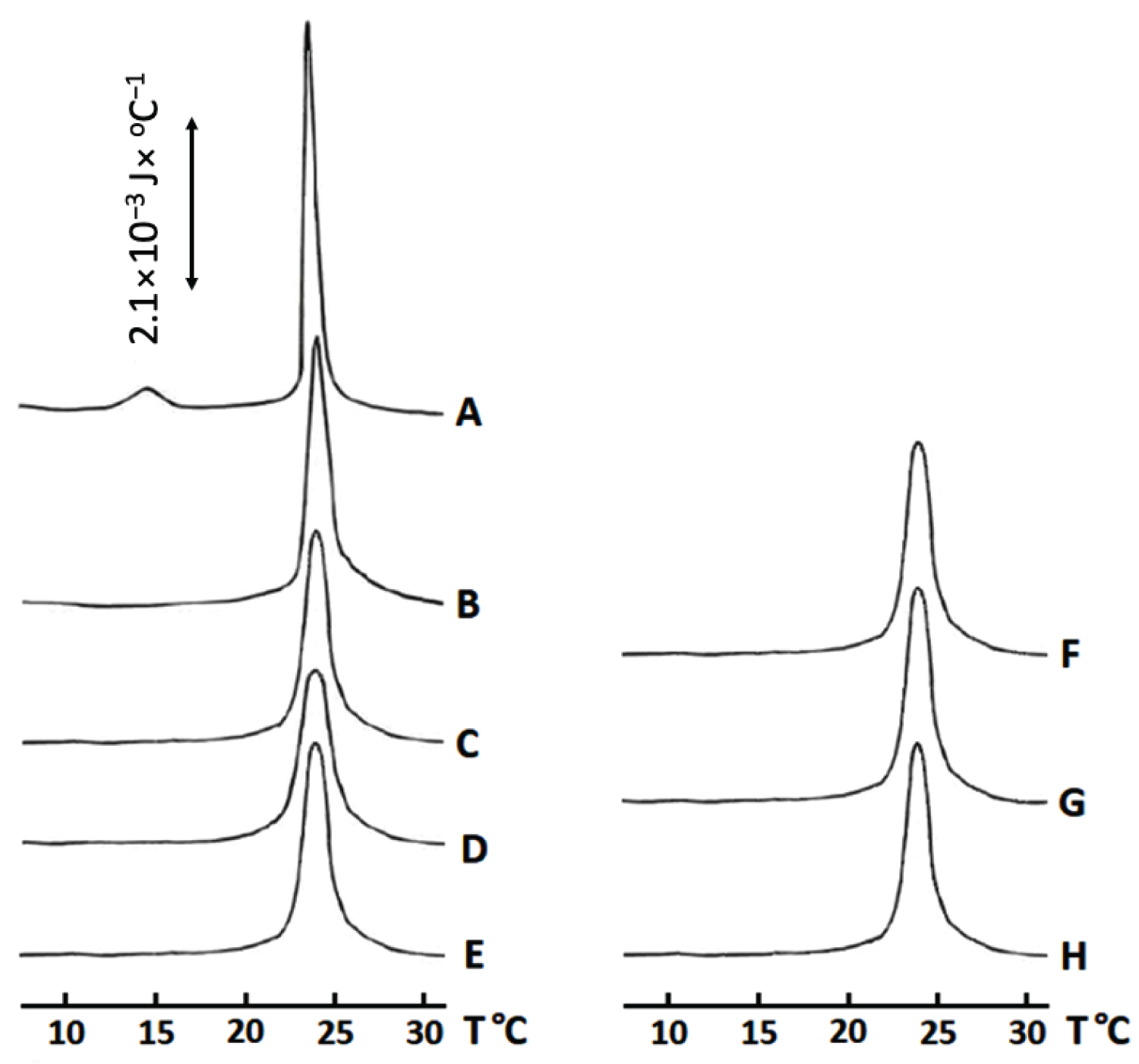
The shape of the calorimetric curve for a saturated lipid phase transition, from a solid crystalline to liquid-gel state, reflects the degree of order in packing of saturated lipids. A calorimetric curve with a narrow main transition peak having an arrow width at peak's half-height (about 1 ℃) and a small pre transition peak located about 10 ℃ below the main transition peak, indicates a high order of packing of saturated lipids [28]. Disturbance in the packing order of saturated lipids in a solid crystalline state by an impurity or by the action of a membrane active protein results in disappearance of pretransition peak, broadening of main transition peak and a decrease in its height and an increase in the width at peak's half-height. Also, when packing order of saturated lipids is disturbed, a recorded enthalpy change of phase transition from solid crystalline to liquid-gel state is decreased. That is because when tight packing of lipids is disturbed, less heat energy is needed to drive the phase transition [28].
The calorimetric curve of DMPC large unilamellar liposomes has a pretransition peak at about 14.5 ℃ and a narrow main transition peak at about 23 ℃ which points to a tight and high order of packing of DMPC molecules in liposomal membranes (Figure 1A). Addition of melittin to DMPC liposomes decreases the value of the enthalpy change, ∆H/∆H0 and broadens the main transition peak at its half-height (Figure 1B and Table 1) making the pretransition peak disappear (Figure 1B). This observation strongly suggests that melittin interacts with DMPC membranes causing a disturbance in packing of DMPC molecules.
Addition of acidic PLA2 to DMPC liposomes resulted in an increased disturbance in the packing order of DMPC molecules compared to addition of melittin alone (Figure 1C and Table 1). This greater disturbance is likely caused, not only by the physical interaction of the acidic PLA2 with DMPC membrane, but also by the esterase activity of the enzyme resulting in the accumulation of lysolipids and fatty acids in liposomal membranes which further disturbs the packing order of lipids in a solid crystalline phase.
Pretreatment of DMPC liposomes with melittin followed by post-treatment with the acidic PLA2 caused a synergistic enhancement in the disturbance of the packing order of DMPC molecules and, additionally, in esterase activity of the acidic PLA2 (Figure 1D and Table 1). This observation suggests that melittin changes the surface of DMPC liposomes making them more susceptible to hydrolysis. Interestingly, when DMPC liposomes were first treated with acidic PLA2 and then post-treated with melittin, both the membrane packing order and esterase activity were reduced compared with DMPC liposomes that were pretreated with melittin and then post-treated with acidic PLA2 (Figure 1E and Table 1). This observation strongly implies that melittin inhibits the esterase activity of PLA2 in DMPC liposomes pretreated with the acidic enzyme.
Unexpected results were obtained with basic PLA2. We anticipated that basic choline moiety will repel basic PLA2 from PC membrane surface to inhibit esterase activity, but that was not the case. Basic PLA2 had a higher esterase activity than the acidic PLA2 and caused more perturbation in membrane packing of the DMPC liposomes (Figure 1C, Figure 1F and Table 1). It is possible that electrostatic attraction of acidic PLA2 to choline groups on membrane surface affected functional conformation of enzyme's active center, while that was not the case with basic PLA2. Melittin inhibited both esterase and membrane disturbing effects of the basic PLA2 on DMPC liposomes regardless of the addition sequence as opposed to acidic PLA2 (Figure 1G, Figure 1H and Table 1). Melittin inhibited the membrane disturbing action and esterase activity of basic PLA2 slightly stronger when melittin was added to the DMPC liposomes following the addition of basic PLA2 (Figure 1G, Figure 1H and Table 1).
Melittin makes a complex with acidic PLA2 at pH 7, but not with basic PLA2
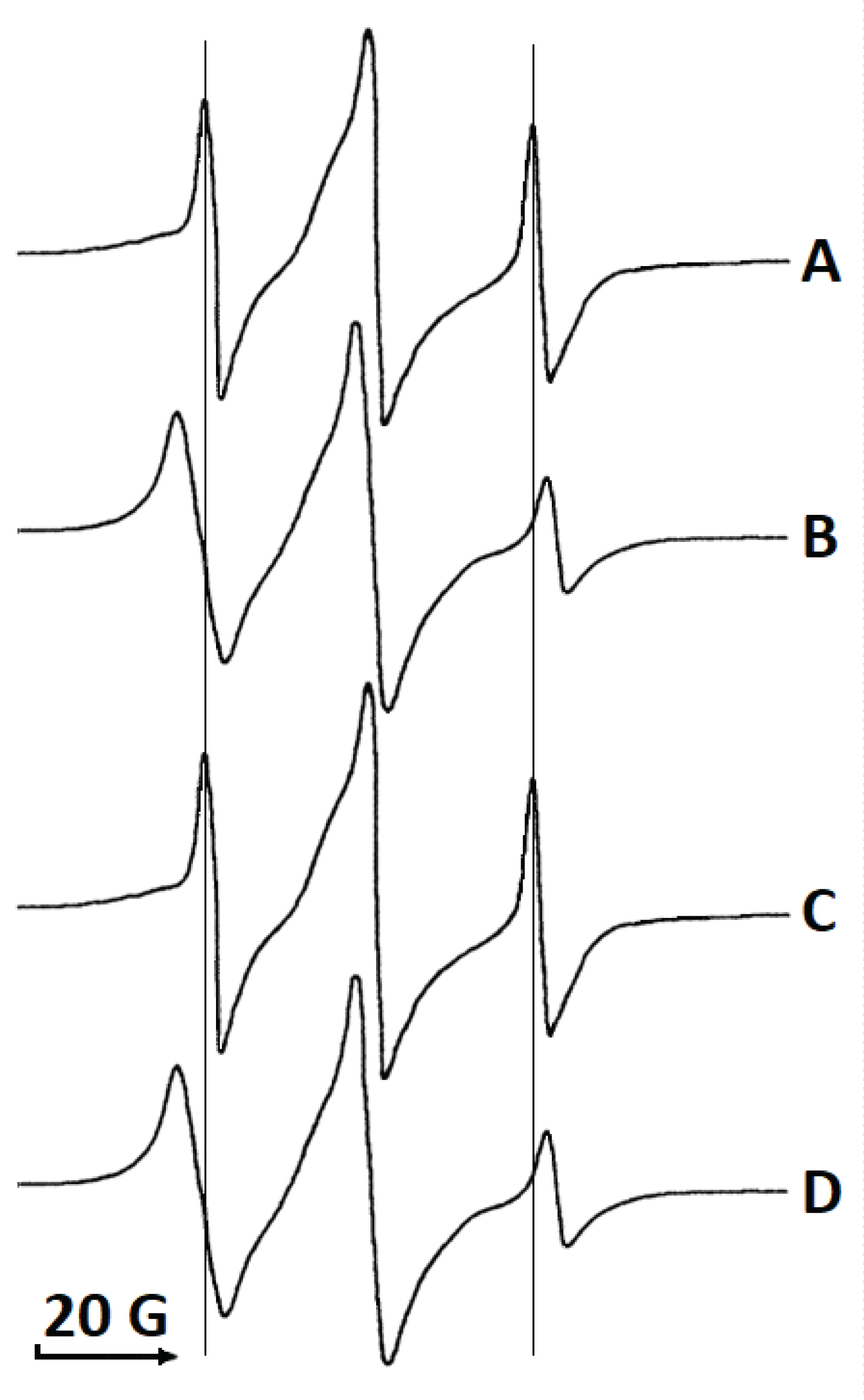
A recent report by Pucca, at al. [22] suggests a mechanism for synergism in cytotoxic actions of PLA2 and melittin in which PLA2 and melittin associate in solution into a complex with increased toxicity. To test whether melittin associates with the acidic or the basic PLA2s in solution we examined the EPR spectra of spin-labeled melittin in buffers of various pHs in the absence or presence of acidic and basic PLA2s. The EPR spectrum of spin-labeled melittin (Figure 2A) appeared as a broader hyperfine splitting with different intensities of resonance peaks than would be expected for a molecule as small as melittin. However, the broadening of the hyperfine splitting with different intensities of resonance peaks results from restricted and axis-symmetrical mobility which aligns well with asymmetrical mobility of melittin that tends to rotate along its long molecular axis. Plus, melittin is known to form dimers and tetramers in solution which may also contribute to the broadening of hyperfine splitting. In addition, dipolar-induced relaxation resulting from the interaction of spin-labels in dimers/tetramers of melittin may contribute to the ERP resonance broadening. The shape of EPR spectrum of spin-labeled melittin in buffer was not affected by changes in pH from 4.0 to 12.5 (data not shown), which may suggest that dimer/tetramerization of melittin in solution is due to hydrophobic forces of attraction, but not forces of electrostatic attraction.
Addition of the acidic PLA2 (pI 4.9) to spin-labeled melittin in buffer at pH 7.0 resulted in a noticeable broadening of the EPR signal (Figure 2B) that most likely comes from association of melittin and the acidic PLA2 molecules, which further restricts the axis-symmetrical mobility of the spin label. Addition of acidic PLA2 to spin-labeled melittin in buffer at pH 4.0 had no effect on the EPR spectrum of spin-labeled melittin (data not shown). This observation strongly suggests that the association of melittin with the acidic PLA2 was electrostatic in nature.
Addition of the basic PLA2 (pI 7.8) to spin-labeled melittin in buffer at pH 7.0 had no affect on the shape of the EPR signal of the spin-labeled melittin (Figure 2C) suggesting that melittin and the basic PLA2 do not associate at pH 7.0. However, when the basic PLA2 was added to spin-labeled melittin in buffer at pH 9.0, the EPR signal of the spin-labeled melittin noticeably broadened (Figure 2D) suggesting that melittin and the basic PLA2 associate electrostatically in a buffer at pH higher than pI value of the basic PLA2.
AutoDock modeling identified ionic and polar intermolecular bonds between melittin and polar head groups of phosphatidylcholine
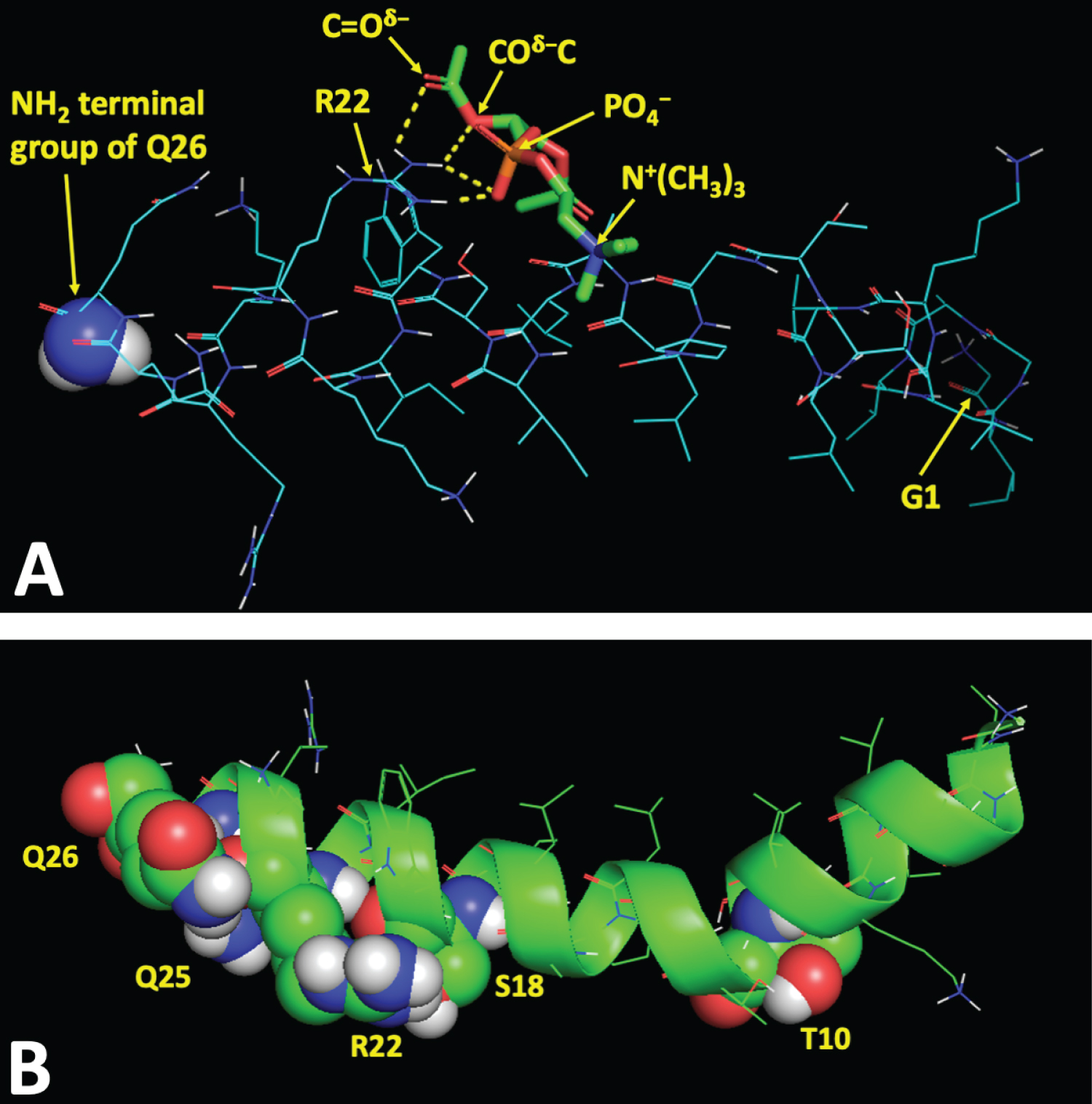
Molecular details of the interaction of amphipathic and cationic melittin with cell membranes, the outer leaflet of which is made of zwitterionic phosphatidylcholine (PC) with basic choline moiety exposed on the outer surface, are poorly understood. To identify the amino acid residues on the molecular surface of melittin, involved in binding to polar headgroups of the PC during melittin interaction with PC membrane, we employed AutoDock Vina software, a powerful program used for docking of macromolecules. In this study, we used only polar head of PC without hydrophobic alkyl chains since alkyl chains are not likely to interact with charged and polar amino acid residues of a membrane-active protein in the polar region of membrane [35]. We analyzed intermolecular bond types between charged and polar amino acid residues of melittin and PC polar headgroups in the nine top binding sites on the molecular surface of melittin with the best binding affinity energies (Table 2). For example, the binding site 1 includes one ionic, one ion-polar, and two hydrogen bonds between PO4-, COδ-C, C=Oδ- polar headgroups of PC and (C=N+H2) charged and (C-NH2δ+) polar groups of amino acid residue R22 of melittin (Table 2). Figure 3A shows in yellow broken lines these four intermolecular bonds between polar head groups of PC and amino acid residue R22 of melittin. The affinity energies varied between top nine binding sites on molecular surface of melittin from -3.50 kcal/mol to -3.20 kcal/mol (Table 2). Overall, only five amino acid residues of melittin, T10, S18, R22, G25 and G26, are involved in intermolecular ionic, ion-polar and hydrogen bonding with polar headgroups of PC (Table 2). All of these five amino acid residues are located on only one side of melittin along its long molecular axis (Figure 3B) suggesting parallel (horizontal) orientation of long melittin axis on the membrane surface with all five residues of melittin, T10, S18, R22, G25 and G26, facing the membrane surface.
Discussion
Melittin, a bee venom membrane-active protein,has been studied extensively [1,3-5] and has a high pharmacological potential for the treatment of various diseases [22]. Melittin binds to membranes composed of PC, but with a higher avidity to membranes enriched with acidic phospholipids [36]. This substrate avidity binding difference explains the higher toxicity that melittin has to cancer cells whose outer cell membranes leaflets are abundant in acidic phosphatidylserine compared to healthy cells which instead contain zwitterionic PC on their outer cell membranes leaflets [36-38]. Our unpublished preliminary studies support this observation: melittin at concentrations of less than 10-5 M exhibits little cytotoxicity to normal human lymphocytes, but at the same concentrations melittin is highly cytotoxic to Jurkat cells, a cell line derived from an acute human T cell leukemia.
Melittin enhances the esterase activity of PLA2 in cancer cells by segregating acidic lipids [36] thereby exposing areas of pure PC and making them more susceptible to attack by PLA2 [37]. Melittin also modulates the esterase activity of endogenous PLA2 in non-cancerous cells, whose outer membrane leaflets are composed of zwitterionic PC [24,37,38]. Melittin thus enhances or mitigates pathophysiological reactions in non-cancerous cells dependent on the levels of hydrolysis of lipid by endogenous PLA2. Melittin is thus a potent anti-cancer agent, but also has promise as a pharmaceutical agent for treating diseases of a non-cancerous nature [2-4,29]. Understanding the mechanisms by which melittin regulates PLA2 activity and the associated physiological reactions is of paramount importance [22].
Two modes of enhancement of PLA2 activity by melittin have been proposed. In one, melittin and PLA2 form a complex whose toxicity is increased to a higher level than the sum of toxicities of each. In the second mode, melittin and PLA2 interact with the same target (membrane phospholipids) resulting in a synergistic enhancement of toxicities of both proteins [39]. In this study we show, by using spin-labeled melittin, that bee venom melittin associates with both acidic and basic PLA2s in buffers at pH values in which both enzymes are negatively charged. This observation strongly implies that melittin associates with PLA2 via electrostatic forces. This also suggests that in vivo melittin will associate with only an acidic PLA2 but not with a basic PLA2.
In contrast to snake venom cytotoxins, which are amphipathic cationic membrane-active proteins that do not bind to pure PC membranes, melittin has a high affinity for the headgroups of PC [24]. By employing differential scanning microcalorimetry assay we showed that melittin binds to DMPC liposomes to disturb molecular packing of lipids. DMPC liposomes pretreated with melittin were more conducive to lipid hydrolysis and further membrane perturbation by acidic PLA2 than DMPC liposomes treated with acidic PLA2 alone. At the same time, when DMPC liposomes were post-treated with melittin, the level of lipid hydrolysis and membrane perturbation was reduced in comparison to that in DMPC liposomes pretreated with melittin. From the estimated amount of fatty acid released by acidic PLA2 (Table 2), a little over 3% of DMPC was hydrolyzed that makes the molar ratio of unhydrolyzed DMPC per melittin about 96 per 1 at a time melittin was added to DMPC liposomes pretreated with acidic PLA2. This means that apart from the acidic PLA2 on the membrane surface to which melittin binds electrostatically, there is still plenty of unhydrolyzed DMPC on the membrane surface for melittin to bind to. Melittin binding to DMPC makes the membrane surface more conducive to lipid hydrolysis by the acidic PLA2. This suggests that the acidic PLA2 in liposomes post-treated with melittin should've produced the same amount of fatty acid that was produced in DMPC liposomes pretreated with melittin. However, that wasn't the case in our study (Table 2), which implies that binding of melittin to acidic PLA2 on membrane surface inhibits esterase activity of acidic PLA2. Thus, two parallel processes, one making membrane surface more conducive for lipid hydrolysis and another inhibiting lipid hydrolysis, produce in overall less amount of free fatty acid than it was produced in DMPC membranes pretreated with melittin. It appears that melittin that first binds to DMPC cannot then bind to acidic PLA2 to inhibit lipid hydrolysis. Overall, the above data suggests that it is the disturbance of DMPC packing on membrane surface caused by melittin binding to DMPC facilitates esterase activity of acidic PLA2, but not the direct interaction (association) of melittin with the acidic PLA2.
Both pretreatment and post-treatment by melittin of DMPC liposomes treated with the basic PLA2 reduced esterase activity of basic PLA2 and the rate of the subsequent membrane perturbation in comparison to that in DMPC liposomes treated with the basic PLA2 alone. This reduction is likely caused by electrostatic repulsion between two cationic proteins, which we have observed in our EPR study in this report. Electrostatic repulsion shields membrane surface area when two cationic proteins compete for the binding on the outer leaflet of DMPC liposomes.
The X-ray analysis of melittin crystals revealed that melittin has a 3D structure of alpha-helical rod that bends at amino acid residue P14 making the 120° angle [33]. As any membrane-active protein, melittin exhibits segregation of the hydrophobic and hydrophilic amino acid residues on molecular surface. The side chains of hydrophobic residues are oriented toward the inside of the bend of the helix, while the side chains of charged and polar residues are oriented toward the outside of the bend [33]. Our AutoDock runs in this study identified five hydrophilic (charged and polar) residues, the side chains of which are oriented toward the outside of the bend of the helix (Figure 3B) suggesting that the outside of the bend of the helix is a molecular site by which melittin binds to PC membrane surface. According to our AutoDock study, R22 with its charged C=N+H2 and polar C-NH2δ+ groups appears to be a key residue driving melittin binding to PC membrane (Table 2). Acidic phosphate group (PO4-) seems to be a key charged moiety of PC polar head that attracts melittin to PC membrane (Table 2). Melittin embeds into polar area of PC membrane with the helical axis parallel to the bilayer plane [33]. Melittin then twists along its long axis to orient hydrophobic side chains inside the bend of the helix toward nonpolar area of alkyl chains in PC membrane. Melittin does not extend to the center of the bilayer and stays in the interface between lipid polar head region and nonpolar area of alkyl chains [33,34]. This location and molecular orientation of melittin inside the lipid bilayer agrees well with circular dichroism and NMR studies with the micelle-bound melittin [40].
One-sided binding of melittin at outer monolayer of PC membrane increases the surface area of outer monolayer over the inner monolayer. A mathematical model calculating changes in membrane shape caused by asymmetric enlargement of monolayer surfaces in the bilayer predicted formation of convexities on the outer surface of membrane [41]. It is plausible that these convexities exposing phospholipids toward solution facilitate esterase activity of PLA2. An increase in one-sided binding of melittin at the outer membrane surface further increases an asymmetrical interfacial area tension. This tension is released by formation of transient pores with melittin helices oriented parallel to the membrane plane [34]. At a lipid to melittin molar ratio exceeding 100 to 1, melittin helices start changing molecular orientation in membrane from parallel to perpendicular to promote formation of stable pores [34]. These changes in membrane structure caused by one-sided binding of melittin at the outer leaflet of membrane may provide multiple opportunities in a structure of lipid substrate interface more conducive to the increased esterase activity of PLA2.
Overall, the results of this study show for the first time by the direct monitoring the order of PC membrane packing and PLA2 esterase activity that melittin significantly increases the acidic PLA2 esterase activity via melittin-induced disturbance of PC membrane parking. We also show for the first time that melittin associates electrostatically with the acidic PLA2 in solution (pH 7) and possibly binds to acidic PLA2 bound to membrane. Our data in this study suggest that direct interaction of melittin with the acidic PLA2 taking place in solution or on membrane surface inhibits esterase activity of enzyme by an unknown mechanism. We have also showed that melittin does not associate with the basic PLA2 in solution (pH 7) and not likely binds to basic PLA2 bound to membrane. Our results in this study suggest that melittin inhibits esterase activity of basic PLA2 as two cationic proteins repel each other and compete for binding to the PC membrane surface. Overall, the results of this study strongly suggest that the melittin-triggered structural changes in a lipid substrate interface, but not a direct interaction of melittin with PLA2, promote synergism in membrane-disintegrating activities of PLA2 and melittin. Finally, the findings of this study warrant further investigation employing site-directed mutagenesis of PC polar head binding sites on melittin surface to corroborate our AutoDock data. In addition, further biophysical studies are needed to elucidate modes of melittin and PLA2 association in solution and on membrane surface to further understand molecular details in mechanism of PLA2 activity modulation by melittin.
Conclusions
The results of this study strongly suggest for the first time that the melittin association with PLA2 enzymes in solution is driven by the forces of electrostatic attraction but not by the hydrophobic forces of attraction. We also suggest for the first time that the melittin binding to the PC membrane surface is presumably propelled by the five amino acid residues, T10, S18, R22, Q25 and Q26, through ionic, ion-polar and hydrogen bonds with the polar headgroups of PC. The C=N+H2 and C-NH2δ+ groups in amino acid residue R22 of melittin and the phosphate group in PC polar head are presumed the key players supporting attraction and binding of melittin to PC membrane. One-side binding of melittin on the outer monolayer of PC membrane not only disintegrates the tight packing of PC polar heads on membrane surface but also creates asymmetrical interfacial tension within a bilayer that promotes transition of lamellar structure to non-lamellar structure including membrane pores. Proposed by us melittin-triggered changes on the PC membrane surface, which are supported by experimental and computer simulation data in this study, make lipid substrate interface more conducive to the esterase activity of PLA2 which is the crucial finding explaining the mechanism of melittin modulation of PLA2 activity on membrane surface. The key amino acid residues identified in this study to promote melittin binding to PC membrane may help with engineering melittin-based anti-cancer drugs and/or antibiotics.
Authors Contribution
Conceptualization, E.S.G.; Data curation E.S.G. and Y.X.; Formal analysis E.S.G. and E.D.R.; Funding acquisition E.D.R. and E.S.G.; Investigation E.S.G., E.D.R., Y.X., P.H.; Methodology E.S.G. and E.D.R.; Project administration E.S.G.; Resources E.S.G. and E.D.R.; Software Y.X. and E.S.G; Supervision E.S.G.; Validation E.S.G. and E.D.R.; Visualization E.D.R. Y.X.; Roles/Writing - original draft E.S.G.; Writing - review & editing E.S.G., E.D.R. and P.H. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
We acknowledge assistance of senior students of the Chaoyang KaiWen Academy in processing AutoDock Vina docking data.
Funding
This research was funded by NIH grants GM08012 and RR08124 (to E.D.R.) and the Poe-Nablo grant for respiratory diseases (to E.D.R.) and by a start-up grant from the Lomonosov Moscow State University (to E.S.G.).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- H Raghuraman, A Chattopadhyay (2007) Melittin: A membrane-active peptide with diverse functions. Biosci Rep 27: 189-223.
- KW Nam, KH Je, JH Lee, et al. (2003) Inhibition of COX-2 activity and proinflammatory cytokines (TNF-alpha and IL-1beta) production by water-soluble sub-fractionated parts from bee (Apis mellifera) venom. Arch Pharm Res 26: 383-388.
- DJ Son, JW Lee, YH Lee, et al. (2007) Therapeutic application of anti-arthritis, pain releasing, and anti-cancer effects of bee venom and its constituent compounds. Pharmacol Ther 115: 246-270.
- LF Leandro, CA Mendes, LA Casemiro, et al. (2015) Antimicrobial activity of apitoxin, melittin and phospholipase A2 of honeybee (Apis mellifera) venom against oral pathogens. An Acad Bras Cienc 87: 147-155.
- PJ Russell, D Hewish, T Carter, et al. (2004) Cytotoxic properties of immunoconjugates containing melittin-like peptide 101 against prostate cancer: In vitro and in vivo studies. Cancer Immunol Immunother 53: 411-421.
- CE Dempsey (1990) The actions of melittin on membranes. Biochim Biophys Acta 1031: 143-161.
- S Ladokhin, ME Selsted, SH White (1997) Sizing membrane pores in lipid vesicles by leakage of co-encapsulated markers: Pore formation by melittin. Biophys J 72: 1762-1766.
- G Carrasquer, M Li, S Yang, et al. (1998) Effect of melittin on PD, resistance and short-circuit current in the frog gastric mucosa. Biochim Biophys Acta 1369: 346-354.
- SW Hui, CM Stewart, RJ Cherry (1023) Electron microscopic observation of the aggregation of membrane proteins in human erythrocyte by melittin. Biochim Biophys Acta 1023: 335-340.
- L Kiesel, T Rabe, G Hauser, et al. (1987) Stimulation of luteinizing hormone release by melittin and phospholipase A2 in rat pituitary cells. Mol Cell Endocrinol 51: 1-6.
- BH Knowles, RW Farndale (1988) Activation of insect cell adenylate cyclase by Bacillus thuringiensis delta-endotoxins and melittin. Toxicity is independent of cyclic AMP. Biochem J 253: 235-241.
- SS Saini, AK Chopra, JW Peterson (1999) Melittin activates endogenous phospholipase D during cytolysis of human monocytic leukemia cells. Toxicon 37: 1605-1619.
- MT Haber, T Fukui, MS Lebowitz, et al. (1991) Activation of phosphoinositide-specific phospholipase C delta from rat liver by polyamines and basic proteins. Arch Biochem Biophys 288: 243-249.
- GB Mahady, C Liu, CW Beecher (1998) Involvement of protein kinase and G proteins in the signal transduction of benzophenanthridine alkaloid biosynthesis. Phytochemistry 48: 93-102.
- SE Mau, H Vilhardt (1997) Cross talk between substance P and melittin-activated cellular signaling pathways in rat lactotroph-enriched cell cultures. J Neurochem 69: 762-772.
- JE Fletcher, MS Jiang (1993) Possible mechanisms of action of cobra snake venom cardiotoxins and bee venom melittin. Toxicon 31: 669-695.
- M Murakami, Y Taketomi, H Sato, et al. (2011) Secreted phospholipase A2 revisited. J Biochem 150: 233-255.
- MD Listei, KB Glasei, RJ Ulevitch, et al. (1989) Inhibition studies on the membrane-associated phospholipase A2 in vitro and prostaglandin E2 production in vivo of the macrophage-like P388D1 cell. Effects of manoalide, 7,7-dimethyl-5,8-eicosadienoic acid, and p-bromophenacyl bromide. J Biol Chem 264: 8520-8528.
- MG Kokotou, D Limnios, A Nikolaou, et al. (2017) Inhibitors of phospholipase A2 and their therapeutic potential: An update on patents (2012-2016). Expert Opin Ther Pat 27: 217-225.
- FF Davidson, EA Dennis (1990) Evolutionary relationships and implications for the regulation of phospholipase A2 from snake venom to human secreted forms. J Mol Evol 31: 228-238.
- LP Vernon, JD Bell (1992) Membrane structure, toxins and phospholipase A2 activity. Pharmacol Ther 54: 269-295.
- MB Pucca, S Ahmadi, FA Cerni, et al. (2020) Unity makes strength: Exploring intraspecies and interspecies toxin synergism between phospholipases A2 and cytotoxins. Front Immunol 11: 611.
- VG Ivkov, GN Berestovskii (1982) The lipid bilayer of biological membranes, Nauka, Moscow.
- G van den Bogaart, JV Guzman, JT Mika, et al. (2008) On the mechanism of pore formation by melittin. J Biol Chem 283: 33854-33857.
- SE Gasanov, ED Rael, M Martinez, et al. (1994) Modulation of phospholipase A2 activity by membrane-active peptides on liposomes of different phospholipid composition. Gen Physiol Biophys 13: 275-286.
- SE Gasanov, MA Alsarraj, NE Gasanov, et al. (1997) Cobra venom cytotoxin free of phospholipase A2 and its effect on model membranes and T leukemia cells. J Membrane Biol 155: 133-142.
- D Deamer, AD Bangham (1976) Large volume liposomes by an ether vaporization method. Biochim Biophys Acta 443: 629-634.
- Ivanov (1988) Calorimetric methods of studying biopolymers and membrane systems. In: Chapter V, A Rubin, Modern methods of biophysical investigations-a practicum of biophysics. (edn), Vyshaya Shkola, Moscow, 203-216.
- DC Wilton (1990) A continuous fluorescence displacement assay for the measurement of phospholipase A2 and other lipases that release long-chain fatty acids. Biochem J 266: 435-439.
- SE Gasanov, NE Gasanov, ED Rael (1995) Phospholipase A2 and cobra venom cytotoxin Vc5 interactions and membrane structure. Gen Physiol Biophys 14: 107-123.
- Trott, AJ Olson (2010) AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455-461.
- MD Hanwell, DE Curtis, DC Lonie, et al. (2012) Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4.
- TC Terwilliger, L Weissman, D Eisenberg (1982) The structure of melittin in the form Icrystals and its implication for melittin's lytic and surface activities. Biophys J 37: 353-361.
- MT Leea, TL Sunc, WC Hungd, et al. (2013) Process of inducing pores in membranes by melittin. PNAS 110: 14243-14248.
- AG Konshina, IA Boldyrev, YN Utkin, et al. (2011) Snake cytotoxins bind to membranes via interactions with phosphatidylserine head groups of lipids. PLoS One 6: e19064.
- TF Aripov, IA Rozenshtein, BA Salakhutdinov, et al. (1987) The influence of cytotoxins from Central Asian cobra venom and melittin from bee venom on the thermodynamic properties of phospholipid bilayer. Gen Physiol Biophys 6: 343-357.
- ML Coleman, EA Sahai, M Yeo, et al. (2001) Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol 3: 339-345.
- M Sebbagh, C Renvoize, J Hamelin, et al. (2001) Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol 3: 346-352.
- H Laustsen (2016) Toxin synergism in snake venoms. Toxin Rev 35: 165-170.
- J Lauterwein, C Bösch, LR Brown, et al. (1979) Physicochemical studies of the protein-lipid interactions in melittin-containing micelles. Biochim Biophys Acta 556: 244-264.
- SE Gasanov, EE Gasanov (1994) An asymmetric enlargement of the monolayer surfaces mechanism of membrane fusion, J Biol Physics 19: 235-242.
Corresponding Author
Edward S Gasanoff, STEM Program, Science Department, Chaoyang KaiWen Academy, A-7, Apt. 611, Yard 46, 3rd Baoquan Street, Chaoyang District, Beijing 100018, China; Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Vorobievy Gory, Moscow 119991, Russia, Tel: +86-18301425873.
Copyright
© 2020 Xu Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.