Predictive Values of Whole Campus Wastewater Surveillance for SARS-CoV-2
Abstract
Screening university wastewater for SARS-CoV-2 emerged as an attractive method to provide ancillary data to bolster detection of viral circulation, as it had the potential to detect the presence of asymptomatic cases or cases that are part of the campus community but not the student population. Our objective was to evaluate the positive and negative predictive values of wastewater screening for COVID-19 cases. We developed and undertook a pilot wastewater screening program for the Fall semester of 2020. Homogenized wastewater influent was collected once per 24 hours, and extracted nucleic acids were interrogated for the presence of SARS-CoV-2 by quantitative reverse-transcriptase PCR. Dates of positive detection were overlayed with dates of confirmed cases in the student population to determine assay sensitivity. SARS-CoV-2 was detected numerous times and was often, but not always, followed by cases of COVID-19. Positive student cases were preceded by detection of SARS-CoV-2 in wastewater by up to 48 hours at each detection. The positive predictive value of wastewater detection for student COVID-19 cases was 0.8105, and the negative predictive value was 0.991. Wastewater screening has the potential to serve as a tool for community-level SARS-CoV-2 surveillance, and is particularly powerful as a negative predictor of disease activity.
Keywords
COVID-19, SARS-CoV-2, Wastewater, Surveillance
Introduction
The novel Betacoronavirus SARS-CoV-2 emerged in late 2019, resulting in a global pandemic of COronaVIrus Disease 2019 (COVID-19) [1]. College campuses were among the many communities tasked with developing methods for surveillance to detect the presence of both positive cases and the virus itself due to its high capacity for horizontal transmission in dense community settings [2-4]. While SARS-CoV-2 is transmitted person-to-person via inhalational spread, the virus can be detected in wastewater. Wastewater screening can therefore be a useful tool to monitor viral presence in communities, as is done for multiple pathogens [5-7]. The University of New England's main campus features a stand-alone wastewater treatment facility serving 1,000 students that is distinct from the surrounding municipal treatment plant, indicating that any detection of SARS-CoV-2 can be definitively linked to the campus community. We utilized this setup to evaluate the positive and negative predictive values (PPV, NPV) of wastewater screening for COVID-19 activity.
Materials and Methods
We developed a wastewater screening program for the Fall semester of 2020. Homogenized influent was collected from the primary settling tank at the wastewater treatment facility once per 24 hours (N = 84 samples).
Wastewater collection
Influent was collected in sterile tubes containing DNA/RNA Shield™ (Zymo Research) at a 4:1 v/v ratio. Organic material was concentrated onto MF-Millipore® hydrophilic mixed cellulose ester 0.45 µm membranes (Sigma-Aldrich) by vacuum filtration [8], removed from the filter surface with sterile cell scrapers, and homogenized in 1 mL fresh DNA/RNA Shield™.
Nucleic acid extraction and molecular testing
Total RNA was extracted using Quick-RNA™ reagents (Zymo Research), and cDNA was generated using qScript™ cDNA Supermix (QuantaBio) per the manufacturer's instructions. Samples were interrogated for the presence of SARS-CoV-2-derived template by quantitative reverse-transcriptase PCR using the TaqPath™ COVID-19 detection platform (ThermoFisher Scientific). Wasterwater samples were considered positive at Ct ≤ 37, as cutoff by Applied Biosystems™ COVID‑19 Interpretive Software for clinical samples. Dates of confirmed diagnoses of COVID-19 in the UNE student community were provided by Student Health Services in the absence of any identifiable personal health information, and used to determine the prevalence of COVID-19 during the Fall 2020 semester (P = 0.049).
Determination of predictive values
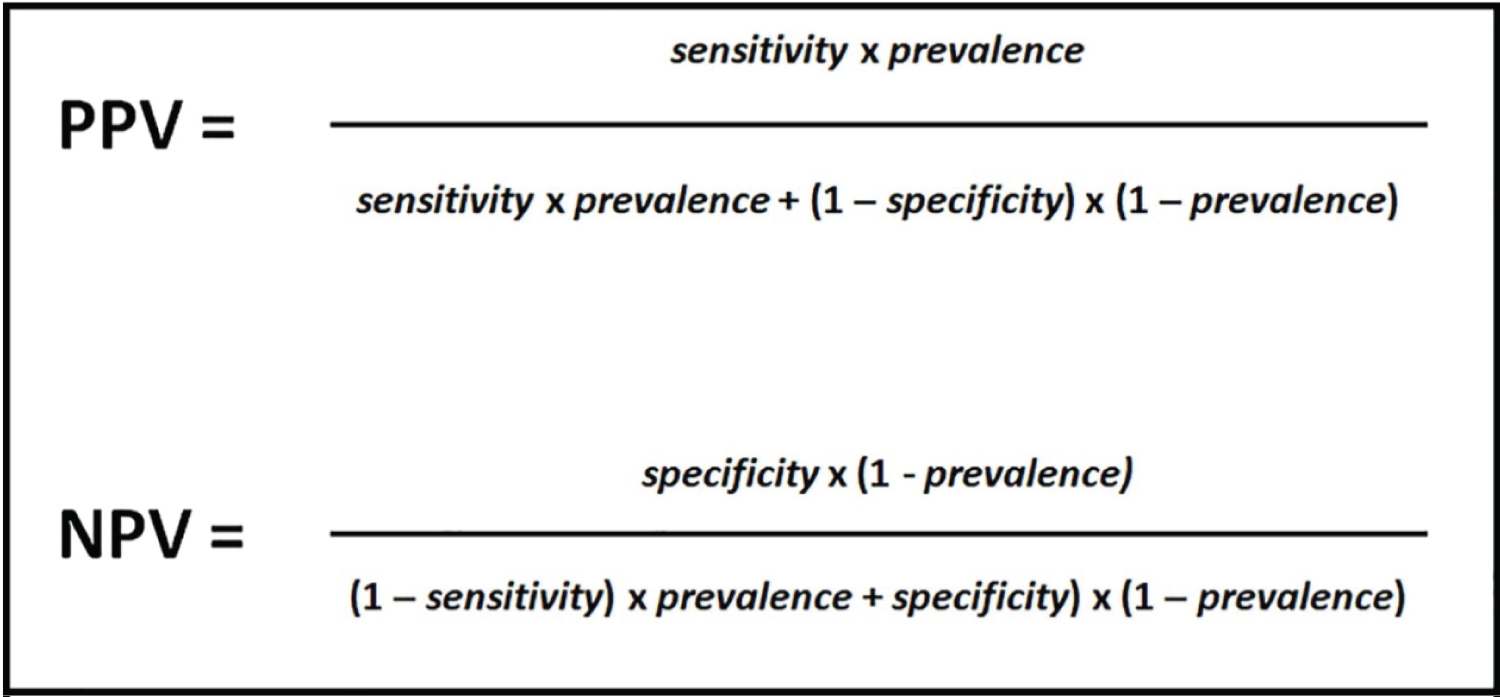
Positive predictive values and negative predictive values were calculated as shown in Figure 1, using specificity value provided for TaqPath reagents and calculating sensitivity by dividing true positives (i.e., confirmed cases) by all positives (i.e., wastewater positives).
Results
SARS-CoV-2 was detected during the Fall 2020 semester. Wastewater detection was often followed by at least one positive student case of COVID-19 within a 48 hour timeframe. The sensitivity of wastewater detection for temporally-associated COVID-19 was 0.83. The PPV of wastewater detection for student COVID-19 cases was 0.8105. All student cases were preceded by detection of SARS-CoV-2 in wastewater by up to 48 hours, and the NPV of wastewater screening for student COVID-19 cases was 0.991.
Discussion
Our preliminary analysis of wastewater screening for SARS-CoV-2 suggests that it is a somewhat robust positive predictor of COVID-19 cases in the student population. It is highly likely that inclusion of confirmed COVID-19 cases among faculty and staff, which were not diagnosed by UNE Student Health Services in the Fall of 2020 due to widespread remote work, and were not included in the prevalence estimation, would enhance the strength of the PPV. Absence of SARS-CoV-2 in campus wastewater was a strong negative predictor of COVID-19 activity, indicating that wastewater monitoring has excellent potential for establishing and maintaining a community as free of SARS-CoV-2 as the pandemic wanes and cases become less frequent.
Acknowledgements
This work was supported by a research grant from the Kahn Family Foundation (MM). We gratefully acknowledge Mr. Tim Baker for his assistance with wastewater homogenate collection.
References
- Guan WJ, Ni ZY, Hu Y, et al. (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382: 1708-1720.
- Hamer DH, White LF, Jenkins HE, et al. (2021) Assessment of a COVID-19 control plan on an urban university campus during a second wave of the pandemic. JAMA Netw Open 4: e2116425.
- Liu Y, Eggo RM, Kucharski AJ (2020) Secondary attack rate and superspreading events for SARS-CoV-2. Lancet 395: e47.
- Ng OT, Marimuthu K, Koh V, et al. (2021) SARS-CoV-2 seroprevalence and transmission risk factors among high-risk close contacts: A retrospective cohort study. Lancet Infect Dis 21: 333-343.
- Alleman MM, Coulliette-Salmond AD, Wilnique P, et al. (2021) Environmental surveillance for polioviruses in Haïti (2017-2019): The dynamic process for the establishment and monitoring of sampling sites. Viruses 13: 505.
- Colosi LM, Barry KE, Kotay SM, et al. (2021) Development of wastewater pooled surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from congregate living settings. Appl Environ Microbiol 2021; 87: e0043321.
- Hendriksen RS, Lukjancenko O, Munk P, et al. (2019) Pathogen surveillance in the informal settlement, Kibera, Kenya, using a metagenomics approach. PLoS One 14: e0222531.
- Ahmed W, Harwood VJ, Gyawali P, et al. (2015) Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl Environ Microbiol 81: 2042-2049.
Corresponding Author
Meghan May, Ph.D., College of Osteopathic Medicine, University of New England, 11 Hills Beach Road, Biddeford, ME, 04005, USA.
Copyright
© 2022 Green E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.