Morphological and Molecular Characterization of Alternaria Solani Isolates Causing Tomato Early Blight in Kenya
Abstract
Early blight (EB) caused by Alternaria solani is among the most devastating tomato diseases in Kenya. In this study, we collected 96 A. solani isolates from three counties in Kenya and characterized them using cultural features, conidial morphology, and genetic variation based on the 18S rRNA gene. Most colonies (45%) were greenish-white in colour with diameter ranging between 65.5-85 mm. Most colonies had concentric zonation (63%) and margins were mostly regular (53%). Conidia was ellipsoidal in most isolates (54%) with lengths ranging between 16.72-20.48 µm. Conidial septa ranged from 2-5 (transverse), 1-3 (longitudinal) and 1-4 (beak septa). Analysis of variance indicated that cultural features and conidial parameters only differed significantly by isolate and not by county of origin. Similarly, the UPGMA dendrogram based on the 18S rRNA gene provided no evidence of geographical clustering of isolates. This has been the first study to characterize A. solani isolates from Kenya. Our results indicate that there is a high level of heterogeneity among Kenya's A. solani populations which will have implications for EB management in the country.
Keywords
Alternaria solani, 18S rRNA, Early blight
Introduction
Alternaria solani (Ellis and G. Martin) Sorauer is a necrotrophic fungus belonging to order Hyphae and family Pleosporaceae [1-3]. The fungus causes Early blight (EB), which is among the most devastating diseases of tomato and potato worldwide. Plants affected by EB usually develop grey-brown lesions on their leaves, leading to reduction in photosynthesizing surface areas and ultimately, yield losses as high as 79% [4].
In Kenya, EB is one of the major biotic constraints of tomato production and causes significant yield losses in farmers' fields. In managing EB, Kenyan farmers have mainly relied on application of fungicides but there have been many complaints recently, about the declining efficacy of most available fungicides [5-7] and this has provided a need, for reinforcing chemical control with alternative strategies, for example, breeding tomato for EB resistance.
However, with A. solani's high polycyclic nature and variability, durable resistance can only be imparted if tomato breeders have sufficient knowledge about the population of the pathogen in Kenya; and which is currently not available. For example, no study has ever determined the level of morphological and genetic variability of A. solani.
The objective of this study was therefore to characterize A. solani isolates from Kenya by colony characteristics, conidial features, and genetic variability (based on 18S rRNA gene). The 18S rRNA gene has been described as highly conserved at intraspecific level [8], so we hypothesized that the variations in it could give a good picture of the extent of intraspecific variation that has taken place in Kenya's A. solani populations.
Materials and Methods
Sample collection
Tomato leaf samples with typical early blight symptoms were collected from farmers' fields in three major tomato-producing counties (Kajiado, Kiambu, and Kajiado) of Kenya during the January-April growing season of 2021. The samples (one per field) were packed in zipper-lock bags and transferred to the Plant Pathology Laboratory at Kenyatta University, Nairobi, Kenya.
Isolation of Alternaria spp
Isolation of Alternaria spp from diseased samples was carried out following the standard protocol by Schulz, et al. [9]. Briefly, each sample was surface-sterilized in 1% NaOCl for 3 min and rinsed three times in sterilized distilled water for 3 min. Sections of ~ 5 mm2 were cut from the advancing edges of the lesions and plated on freshly prepared PDA media, the petri plates sealed and incubated at 27 °C for 5 days. Subculturing was done for A. solani putative colonies [10], continuously until pure cultures were obtained.
Cultural and morphological characterization of isolates
This was done on 3 replicate cultures of each isolate at 7 days. The characteristics recorded include colony diameter, colour, nature of margin and colony zonation [11]. When the cultures were 3 weeks old, morphological characterization was carried out, following methods by previous workers [11-13].
Using a sterile scalpel blade, small sections of hyphal tips were 'brushed' onto a 1ml drop of sterile distilled water, on a rectangular slide. Such slides were examined under a Zeiss - Primo Star microscope fitted with an AxioCam ERC 5s camera at 40X. The features recorded include conidial parameters (such as shape, length, and width) and number of septa. For analysis, quantitative variables (such as colony diameters and conidia lengths) were subjected to one-way ANOVA while qualitative variables such as colony colours and nature of margins were summarized in form of frequencies and percentages.
Pathogenicity tests
Conidial suspensions were prepared from 20 randomly selected isolates according to Stammler, et al. [14] and inoculated onto leaves of three-week-old tomato seedlings of Cultivar 'Riogrande' (3 replicates per isolate). For the control experiment, seedlings were inoculated with sterile distilled water. These tests were conducted under greenhouse conditions in pot experiments. Leaves showing typical early blight symptoms were collected from infected plants, two weeks after inoculation and the pathogen re-isolated to complete Koch's postulates.
DNA extraction and PCR amplification of the 18S rRNA gene
DNA was extracted from mycelial cultures using a Qiagen DNA extraction kit (Qiagen GmbH, Germany) as per the manufacturer's protocol. Two microlitres of purified DNA per isolate were used in 50 µl of PCR mixture.
The primers used (5'-CACAAGGACCAACCCATAAAC-3'(forward) and 5'-CGCAAGGGGAGACAAAAAG-3' (reverse), were sourced from Macrogen Inc, Netherlands. Thirty microlitre PCR volumes contained 1.5 µL of 10 µm/µL forward primer, 1.5 µL of 10 µm/µL reverse primer, 15 µL of Taq DNA polymerase, 2.0 µL template DNA, and 10 µL nuclease-free water. PCR was conducted in a gradient thermal cycler (Applied Biosystems) and involved four stages; initial denaturation at 94 °C for 5 min, 25 cycles of extension at 94 °C for 1 min, annealing at 58 °C for 1 min, and final extension at 72 °C for 5min.
To confirm amplification, two microlitres of each PCR product were electrophoresed through 1% agarose gel at 100 V for 30 min. A two kilobase DNA ladder was used as a size standard. After electrophoresis, the gels were visualized under a UV transilluminator. PCR products were then cleaned by ethanol precipitation [15] and sent to Macrogen Inc. (Netherlands) for sequencing.
Bioinformatics analysis of the sequences
The sequencing quality of reads was assessed visually using the Bioedit® software whereby, overlapping sections (noise) were trimmed off the chromatograms. To support morphological identification of isolates, trimmed DNA sequences were analyzed for homology with published Alternaria solani sequences using NCBI's Blastntool
(https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). Phylogenetic analysis was then performed in MEGA7 software [16] by construction of a phylogenetic tree (using UPGMA method) to resolve relationships among the isolates.
Results and Discussion
Cultural characterization
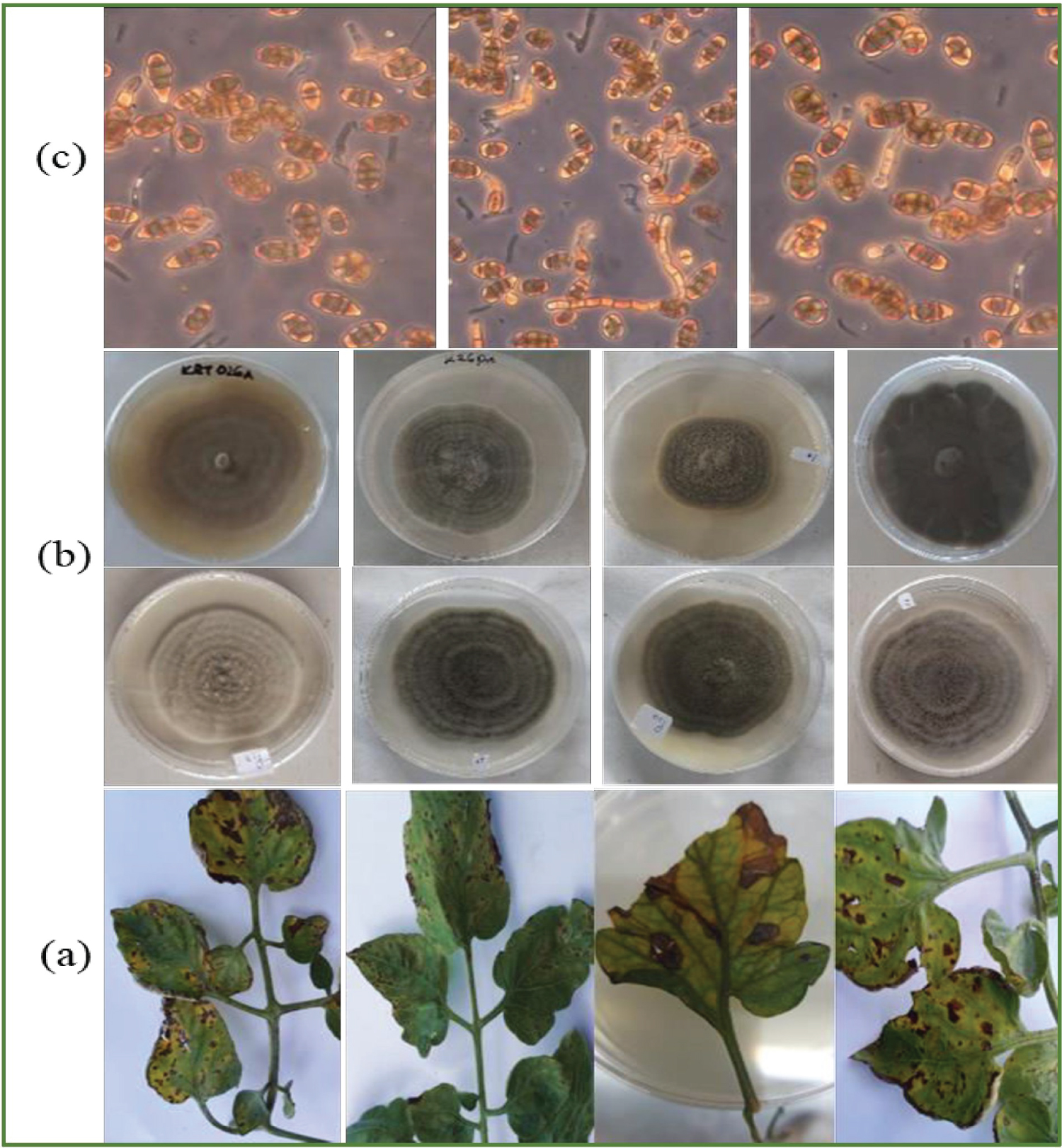
Initially, the mycelia in all cultures were profuse and hyaline but turned grey to brown, septate, and branching as they grew. Isolates exhibited significant variation in cultural features (colony diameter (P = 0.031), colony colour (P = 0.029)) though this variation was not significant when isolates were grouped by county of origin (Table 1). Isolate KYG24 from Kirinyaga had the highest recorded colony diameter at 85 mm while KJD18 from Kajiado had the lowest colony diameter at 65.5 mm.
Regarding colour, most colonies (45 of 96 or 45%) were greenish-white, 22 were creamish-white, 10 green and 1 grey. On reverse plate, most colonies were pigmented creamish white (49 out of 96), greenish-brown (42) and brown (5). About half (55.2%) of the isolates had irregular margins and majority (62.5%) of isolates had concentric zonation (Figure 1). These results are in agreement with previous studies about the high variability in cultural features among A. solani isolates. However, since cultural characteristics in fungi are often affected by many factors e.g media used and incubation temperature [17], we only used them for initial screening of Alternaria spp and did not infer much about them for characterization of the pathogen.
Morphological characterization
In all cultures, hyphae were hyaline and relatively thick (4.41 - 4.44 µm diameter). Conidiophores were 200-230 µm, flexuous or straight, and were either solitary in many isolates or in small groups in a few others. At apices, conidiophores enlarged slightly with scars indicating points of conidia attachment. Conidia were pale to olivaceous-brown, borne singly or in short chains, on conidiophores. They were straight, or slightly flexuous tapering to a beak in most isolates or flat-ended in others.
Conidia lengths ranged from 16.72-20.48 µm (Mean 18.46, St. dev. 3.83, P = 0.031) while widths were between 11.87-12.13 µm (mean 11.44, St. dev.2.2, P = 0.028) (Table 2). Three conidia shapes were identified; ellipsoidal (54%), obclavate (35%), obvoid (10%). All conidia had at least 2 transverse septations (range 2-5). Majority (69.8% or 67 of all isolates) had longitudinal septa in their conidia, ranging from 1-3. Only 56% (or 56 isolates) had beaked conidia. Beak lengths ranged from 19.7-6.8 µm (Mean 10.9, St. dev. 3.8, P = 0.035) while the number of beak septa varied from 1-4.
These results indicate a high level of morphological variability among Kenya's A. solani isolates as has been reported in other studies [12,18,19]. However, this variation was not significant at county of origin level, which confirmed existence of morphological heterogeneity among A. solani isolates even when they were isolated from the same geographical area/agroclimatic zone.
Pathogenicity tests for Alternaria solani isolates
Symptoms started to appear 3 days after spraying the A. solani inoculations. Brown, irregular spots (2-4 mm in diameter) with concentric zonations at the center, appeared on leaves. In some cases, the spots enlarged in size reaching up to 10 mm in diameter in the second week after inoculation. Re-isolated cultures from infected leaves had close similarity with inoculated A. solani isolates in terms of cultural and morphological features. This confirmed the pathogenicity of the tested isolates on tomato as per Koch's postulates.
Molecular characterization
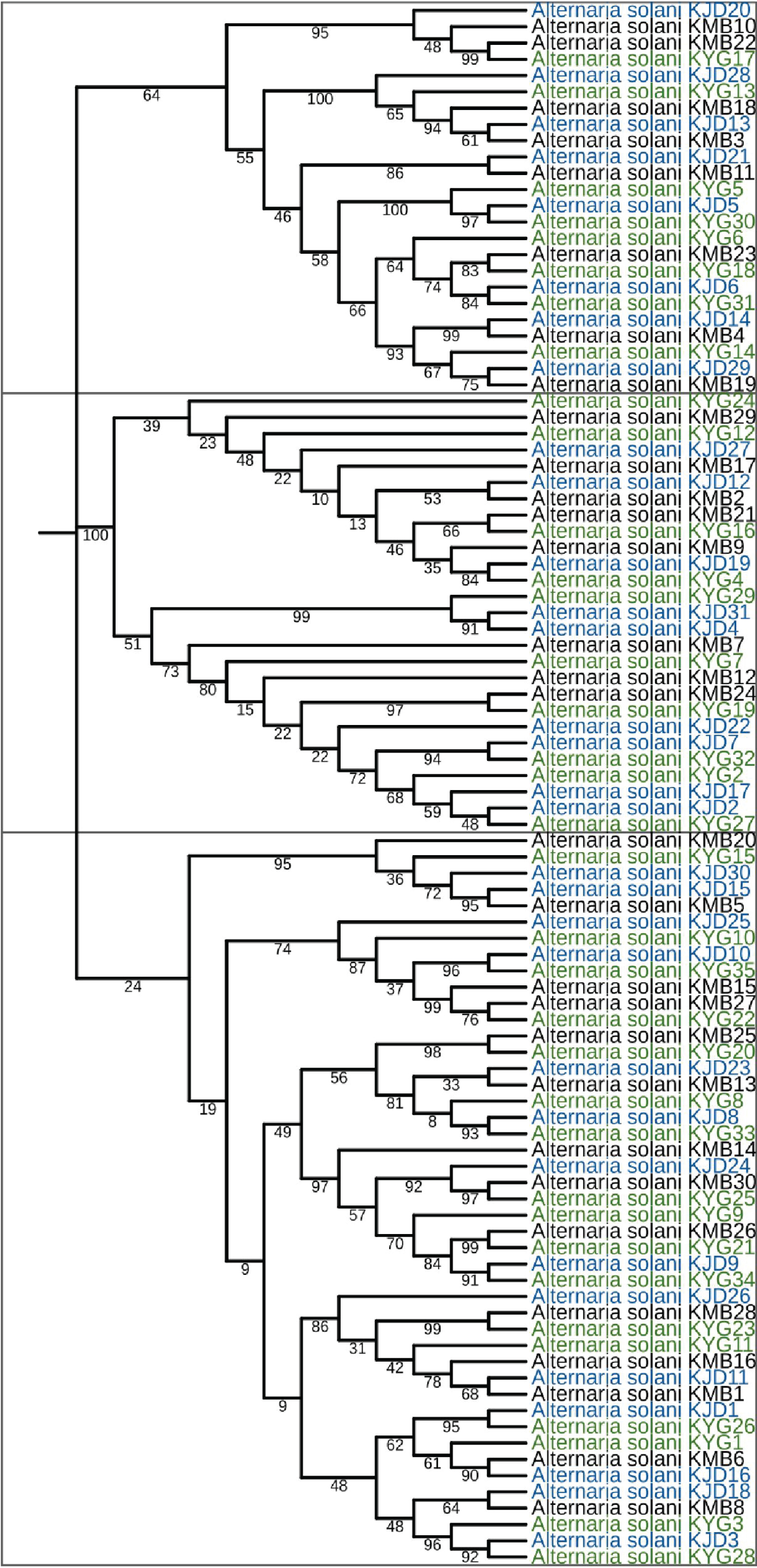
The two primer pairs amplified an ~ 1600 bp fragment of 18S rRNA gene in each A. solani isolate. Trimmed DNA sequences, ~ 1500 bp long were obtained for Kirinyaga (n = 35), Kiambu (30), and Kajiado (31) counties. In the GenBank database, sequences showed high percent similarities (98-99.5%) with Alternaria solani accession No. KC478609.1. This served as the final confirmatory step for Alternaria solani.
During phylogenetic analysis, three distinct clusters (containing 24, 27, and 45 isolates, respectively) were formed (Figure 2). In agreement with previous studies in other areas [20,21], the isolates in the present study did not cluster according to the county of origin.
This finding confirms presence of genetically heterogeneous A. solani isolates in similar geographical regions in Kenya and may mean that the pathogen population in the country has a high adaptive potential to its environment. The same trend has been reported in A. solani populations in other countries such as Germany [22], India [23], and Brazil [24]. With this genetic variability, A. solani in Kenya may easily challenge EB management programs in Kenya by evolving resistance to fungicides and/or overcoming host resistance to early blight. Continuous monitoring of A. solani populations in Kenya especially through studies on pathogenic variability and sensitivity to fungicides will therefore provide an important knowledge base for improvement of EB management practices in the country.
Conclusion
This study characterized Alternaria solani isolates from tomato in Kenya and revealed that the isolates differed significantly in cultural characteristics, morphological features, and phylogenetically, based on the 18S rRNA gene. This finding will have implications for Early bight management in Kenya.
Data Availability Statement
All data supporting the findings of this study has been included in the manuscript and its supporting files.
Acknowledgments
The authors are grateful to the agriculture departments of Kirinyaga, Kiambu, and Kajiado counties for collaboration during sample collection. This study received funding from the National Research Fund, Kenya (Grant title: ENABLE-HORT) and the authors appreciate this support.
Competing Interests
The authors declare that there were no competing interests.
Author Contributions
M.M and S.R conceptualized the study and obtained funding. A.M.N collected the data and wrote the draft manuscript under guidance by M.M and S.R. All authors read and approved the final manuscript for publication
Supplementary File 1
Supplementary File 2
References
- Ghafri AA, Maharachchikumbura SSN, Hyde KD, et al. (2019) A new section and a new species of Alternaria encountered from Oman. Phytotaxa 405: 279-289.
- Lawrence DP, Rotondo F, Gannibal PB (2016) Biodiversity and taxonomy of the pleomorphic genus. Alternaria. Mycol Prog 15: 1-12.
- Stuart RM, Bastianel M, De Azevedo FA, et al. (2009) Alternaria brown spot. Fitopatol 30: 29-44.
- Foolad MR, Merk HL, Ashrafi H (2008) Genetics, genomics and breeding for late blight and early blight resistance in tomato. Plant Sci 27: 75-107.
- Mugao LG, Muturi PW, Gichimu BM, et al. (2020) In Vitro control of phytophthora infestans and Alternaria solani using crude extracts and essential oils from selected plants. Int J Agron 2020: 1-10.
- PCPB (2019) Annual report 2019 Pesticides Control and Products Board. Nairobi, Kenya.
- Mwangi MW, Kimenju JW, Narla RD, et al. (2015) Tomato management practices and diseases occurrence in Kirinyaga West Sub County. J Nat Sci Res 5: 119-124.
- Wu S, Xiong J, Yu Y (2015) Taxonomic resolutions based on 18S rRNA genes: A case study of subclass Copepoda. PLOS one.
- Schulz B, Wanke U, Draeger S, et al. (1993) Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol Res 97: 1447-1450.
- Chohan S, Perveen R, Abid M, et al. (2015) Morpho-physiological studies management and screening of tomato germplasm against Alternaria solani the causal agent of tomato early blight. Int J Agric Biol 17: 111-118.
- Marak RT, Ambesh BS, Das S (2014) Cultural, morphological and biochemical variations of Alternaria solani causing diseases on solanaceous crops. Bioscan 9: 1295-1300.
- Nikam PS, Suryawanshi AP, Chavan AA (2015) Pathogenic, cultural, morphological and molecular variability among eight isolates of Alternaria solani, causing early blight of tomato. Afr J Biotechnol 14: 872-877.
- Kumar V, Singh G, Tyagi A (2017) Evaluation of different fungicides against Alternaria leaf blight of tomato (Alternaria solani). Int J Curr Microbiol App Sci 6: 2343-2350.
- Stammler G, Bohme F, Philippi J, et al. (2014) Pathogenicity of Alternaria-species on potatoes and tomatoes. Fourteenth Euroblight Workshop PPO-Special Report 16: 85-96.
- Green MR, Sambrook J (2016) Precipitation of DNA with Ethanol. Cold Spring Harb Protoc.
- Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 38: 3022-3027.
- Slepecky RA, Starmer WT (2009) Phenotypic plasticity in fungi: A review with observations on Aureobasidium pullulans. Mycologia 101: 823-832.
- Loganathan M, Venkataravanappa V, Saha S, et al. (2016) Morphological, pathogenic and molecular characterizations of Alternaria species causing early blight of tomato in Northern India. Proc Natl Acad Sci Indian Sect B Biol sci 86: 325-330.
- Naik MK, Prasad Y, Bhat KV, et al. (2010) Morphological, physiological, pathogenic and molecular variability among isolates of Alternaria solani from tomato. Indian Phytopathol 63: 168-173.
- Husnain L, AlKahtani M (2019) Molecular heterogeneity in the 18s DNA gene of Alternaria sp. and Fusarium sp. producing mycotoxins in rice and maize grains. Saudi J Biol Sci 26: 368-372.
- Ismail AWA, Sidkey NM, Arafa RA, et al. (2016) Evaluation of in vitro antifungal activity of silver and selenium nanoparticles against Alternaria solani caused early blight disease on potato. Biotechnol J Int 12: 1-11.
- Leiminger JH, Auinger HJ, Wenig M, et al. (2016) Genetic variability among Alternaria solani isolates from potatoes in Southern Germany based on RAPD-profiles. J Plant Dis Prot 120: 164-172.
- Varma PK, Singh S, Gandhi SK, et al. (2006) Variability among Alternaria solani isolates associated with early blight of tomato. Commun Agric Appl Biol Sci 71: 37-46.
- Lourenço Jr V, Rodrigues TT, Campos AM, et al. (2011) Genetic structure of the population of Alternaria solani in Brazil. J Phytopathol 159: 233-240.
Corresponding Author
Andrew M Nuwamanya, Department of Agricultural Science and Technology, School of Agriculture and Enterprise Development, Kenyatta University, PO Box 43844, Nairobi, Kenya
Copyright
© 2022 Nuwamanya AM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.