Physiological Races of Colletotrichum lindemuthianum from South and Central Ethiopia
Abstract
Bean anthracnose (Colletotrichum lindemuthianum) is one of the major diseases of common bean (Phaseolus vulgaris) in Ethiopia causing up to 63% yield loss. The current study was initiated with the objective to identify races of C. lindemuthianum prevailing in major bean producing areas in south and central Ethiopia. Thirty isolates of C. lindemuthianum, collected from Damot Gale, Halaba Special, Adama, Boset, Hawassa Zuria and Boricha woredas in 2017 were inoculated on 12 differential cultivars in greenhouse using Completely Randomized Design (CRD). Results revealed the presence of 17 physiological races (pathotypes), of which only three were previously reported from Ethiopia. Race 9 was the most dominant race across the surveyed areas. Four of the 17 races (3047, 2260, 2225 and 2073) were able to infect the highly resistant differential cultivar G2333 indicating that the Ethiopian C. lindemuthianum populations might be composed of highly virulent races. The differential cultivars Michelite, Mexico 222 and PI 207262 showed the most susceptible reaction to the tested races, while no differential cultivar was immune to the pathogen races. In conclusion, the current work demonstrated the existence of highly variable isolates of C. lindemuthianum that incite bean anthracnose in Ethiopia. The generated results of race identification would serve as important inputs for future development of host resistance against bean anthracnose in Ethiopia and elsewhere having similar agro-ecologies. Therefore, for the future race identification study need to be continued with more advanced techniques and by including more isolates across agro-ecology gradients.
Keywords
Bean anthracnose, Colletotrichum lindemuthianum, Differential cultivars, Phaseolus vulgaris, Physiological races, Ethiopia
Introduction
Bean anthracnose caused by Colletotrichum lindemuthianum (Sacc & Magnus) Briosi & Cavara, is one of the most important seed borne diseases of common bean in the world [1-3]. Depending on the cultivar and environmental conditions, anthracnose infections can drastically reduce crop yield [4]. Fernandez, et al. [5] reported that infection of susceptible cultivars, like Mexican-142 and Awash-1, under favorable environmental conditions leads to 100% grain yield loss. In Africa, the disease is common in Burundi, Democratic Republic of Congo, Ethiopia, Kenya, Rwanda, Tanzania and Uganda [6]. In Ethiopia it is a major production constraint in bean-growing regions [3], causing a yield loss of up to 63% [7].
Colletotrichum lindemuthianum exists in many physiological forms in all bean-growing regions of the world and infects bean genotypes from both bean gene pools, i.e. Andean and Mesoamerican (Mahuku and Riascos, 2004) [8]. According to CIAT [9], the fungus is known to have races that vary across countries, regions, locations and varieties. The highest diversity and variation are reported from Latin America, which is the center of origin of common beans [10]. Since east African regions are considered as the secondary center of diversity for common bean, due to co-evolution of the C. lindemuthianum pathogen and its host, the East African highland regions are expected to have a high variability of C. lindemuthianum [11]. The existence of high pathogenic variability and emergence of new pathogen races results in continuous breakdown of host resistance [12].
Knowledge on race variability of the pathogen population is a prerequisite for developing durable resistance in bean varieties [6]. Co-evolution of the pathogen and its host in the Andean and Mesoamerican gene pools provides a useful means of identifying appropriate sources of resistance, since common bean genotypes originating from one gene pool are more likely to express resistance to pathogenic races than another gene pool [13,14]. In Ethiopia, some efforts have been done to identify the type of races that occur in Ethiopia where Tesfaye (2003) gave an account of eight races of Colletotrichum lindemuthianum from Ethiopia and compared them to six races brought from Southern Africa. But as previous studies in this topic are very limited in Ethiopia, it is important to first characterize the Colletotrichum lindemuthianum isolates collected from major bean growing areas of the country to develop resistant common bean genotypes. Therefore, the current study aimed at filling the gap by assessing race variability among the Colletotrichum lindemuthianum populations of south and central Ethiopia.
Materials and Methods
Disease survey
A field survey for Colletotrichum lindemuthianum isolate collection was conducted in 2017 main season in two regions (Oromia and SNNPR) at six districts, i.e., Damot Gale, Halaba Special (Choroko 2 kebele), Adama (Melkassa), Boset (Kechachula), Hawassa Zuria (Hawassa Agricultural Research Center) and Boricha (Figure 1). The survey districts were systematically selected based on production potential and previous reports on bean anthracnose. Two to three farmers' fields were randomly identified per district for the field disease survey and isolate collection. Five sampling quadrats (1 m2) were used on each farmer's field to collect data on anthracnose incidence and severity as follows:
Disease Incidence (DI) =
Severity of bean anthracnose was recorded on leaves based on CIAT (1987), a standard system for the evaluation of bean germplasm (Table 1).
Sample collection
In each location, samples depicting typical bean anthracnose symptoms were randomly considered, and from each identified representative plants, three leaves were collected. Collected leaves were immediately wrapped on paper towels to absorb their moisture, placed in paper bags and labeled with pencil. Finally, three representative samples from a single common bean leaf with typical anthracnose symptoms from each field were picked for race analysis.
Isolation of the pathogen
Isolation of the pathogen was conducted at Melkassa Agricultural Research Center (MARC) Plant Pathology Laboratory. Leaf samples were washed in running tap water and leaf pieces or patches with 2-5 mm size were cut with sterilized pair of scissors and surface-sterilized with 2.5% sodium hypochlorite (chlorox, NaOCl) solution for 2-3 minutes and rinsed three times with sterile distilled water (SDW). Since collected samples could possibly carry more than one strain of a pathogen or even more pathogens, effort was made to spatially resolute a spot with typical anthracnose symptom from a single leaf, and then pieces were cut out to maintain the genetic purity of Colletotrichum lindemuthianum isolates. Then, the pieces were placed on sterilized paper towel for 5-10 minutes; and five such pieces were aseptically transferred into a 9 cm Petri dish with potato dextrose agar (PDA). The cultures were incubated at 20 ℃ under continuous fluorescent light for 10 days. Sub-culturing was done by taking a 3 mm mycelial plug from the PDA, which was visually free from any contamination, and transferring into new Petri dishes containing fresh PDA. Then continuous sub-culturing was made until pure cultures with no contamination were obtained. While culturing, streptomycin sulfate (5 mg L-1 of PDA) was used to suppress bacterial colonial growth. In this way, a total of 30 isolates were purified and obtained from the survey areas for race identification.
Inoculum preparation for race identification
Two-week-old pure cultures were flooded with 50 mL of sterile distilled water (SDW) and the spores were scraped off using a fine brush. Flooding was repeated three times, each time using fresh sterile distilled water to get most of the conidia from the culture. Then the suspension was poured into a beaker and mixed thoroughly. Spore suspensions from all cultures of the same isolate were mixed and passed through double layer cheesecloth (44*36 thread/sq. inches) to retain fragments of mycelia and culture medium. The concentration of the spore suspension was adjusted to 1.2 × 106 spores per mL of sterile distilled water using hemocytometer. One drop of Tween 20 was added per 100 mL spore suspension and mixed thoroughly before inoculation.
Planting common bean differentials
Common bean seeds of 12 differential cultivars were surface sterilized with 0.1% chlorox solution for three minutes, washed or rinsed thoroughly with sterile distilled water and dried at room temperature (25 ℃ ± 2). Then the seeds were sown in plastic pot filled with 3 kg mix of sun-dried sterile topsoil, manure and sand in 2:1:1 ratio, respectively, and pots placed in greenhouse. Five seeds per pot were sown in completely randomized design, and each plant was considered as a replicate for each common bean differential cultivars and for each isolate of the pathogen.
Inoculation
Fourteen-day-old common bean seedlings were used for inoculation. All the 12 differential cultivars were inoculated separately with the prepared separate isolate suspension (60 × 106 spores per pot) using a hand sprayer on both the abaxial and adaxial surfaces of the leaves until suspension runoff at a time. A control (plants inoculated with sterile distilled water alone) was included for each set of the differentials inoculated with an isolate. Inoculated plants were covered with transparent polythene sheets to maintain high relative humidity. The polythene sheet was removed five days after inoculation. After removal of polythene sheet, pots were kept in the greenhouse and maintained at temperature of 18-33 ℃ and relative humidity ranging from 72 to 95%.
Race determination
The reaction [susceptible reaction (+) and resistant reaction (-)] of each common bean differential cultivar to each isolate was assessed 15 days after inoculation, i.e., 30 days after planting. For the purpose of consistency, only the primary leaves of each plant were evaluated. After evaluation of all differential cultivars, each isolate was assigned a name (race number) based on the binary nomenclature system by adding the numerical binary values of the susceptible varieties together. A binary number is equal to 2n, where n is equivalent to the place of the cultivar within the differential series order as tabulated below (Table 2). The sum of all binary numbers of cultivars with susceptible reactions (i.e., y = 2n + 2(n+1) +2(n+2) +....... +2(n+11); where n = 0) gives a specific race number or name (y) [15].
Results and Discussion
Disease Survey
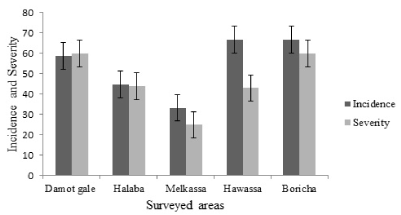
Surveyed areas had elevation ranging between 1436 and 1958 m.a.s.l. Bean anthracnose incidence in the study areas ranged from 33 to 67% (Figure 2). The highest anthracnose incidence (66.7%) was recorded in Hawassa and Boricha, followed by Damot Gale (58.5%) and the lowest (33.3%) was in Melkassa. These results are in contrast to the finding of Habtu, et al. [16] who reported high level of bean anthracnose in the Rift Valley (Melkassa) and low level of bean anthracnose in Sidama (Hawassa).
The mean anthracnose severity score across the surveyed areas ranged from 25 to 60%. The highest (60%) severity score was recorded in Damot Gale and Boricha areas and the lowest (25%) at Melkassa. These results were not in agreement with the observation of Tesfaye [17] who reported higher anthracnose severity in the western zone and Rift Valley areas of the country, suggesting a possible shift in bean anthracnose intensity across the country. Seed born nature of the disease [18] and uncontrolled exchange of common bean seeds for planting across the country lead to such deviation on the incidence and severity of bean anthracnose.
Race determination
The reaction of bean differential cultivars to selected isolates was variable. Out of the 30 isolates of Colletotrichum lindemuthianum collected, a total of 17 physiological races (pathotypes) were identified (Table 3). Race designation gives an indication of the variability of the pathogen [6]. Out of 17 races identified by the current study, only three races, i.e., race 1011 at Areka, race 898 at Bako, and race 128 at Ziway, were previously reported in Ethiopia [17].
Race 9, which was identified from Boricha, Hawassa and Halaba districts, was the most dominant physiological race, followed by the typical races 2260, 272 and 1011. Similar result was reported by Ansari, et al. [19] who reported race 9 as the most widespread race in Argentina. The same authors indicated that race 9 included isolates originating from four different countries. On the other hand, Gonçalves, et al. [20] stated that race 73 was the most common and widespread race in Santa Catarina State in Brazil.
When C. lindemuthianum races identified in the current work are assigned to geographic region, four races (385, 3047, 9 and 587) were from Boricha; two races (2260 and 272) from Melkassa; 12 races (272, 321, 9, 1172, 898, 128, 465, 73, 1250, 34, 1011 and 2260) from Hawassa; two races (2225 and 1011) from Wolayta; and two races (2073 and 9) were from Halaba. This suggests the richness of the Rift Valley region with reference to C. lindemuthianum race variability. The differences among the pathogen populations in different areas might reflect the differences in the agricultural practices employed and common bean germplasm used; that would affect the selection and adaptation processes, in the different agro-ecologies [6].
Of the 17 C. lindemuthianum races identified in the current work, four races, i.e. 3047, 2260, 2225 and 2073, were able to infect the highly resistant differential cultivar G2333. Susceptibility of this cultivar to isolates of C. lindemuthianum has rarely been reported [21]. Thus, the result strongly suggested that C. lindemuthianum populations in Ethiopia could possibly be composed of highly virulent races that may cause much damage even to resistant/tolerant germplasms. Besides causing bean anthracnose on G2333, race 3047 also successfully infected the differential cultivars TU, TO, PI 207262, Mexico 222, Kaboon, Perry Marrow, MDRK and Michelite. Furthermore, race 2260, 2225 and 2073 managed to break the resistance of the differential cultivars PI 207262, Mexico 222, Widusa, Perry Marrow, Kaboon, Cornell 49-242 and Michelite (Table 3).
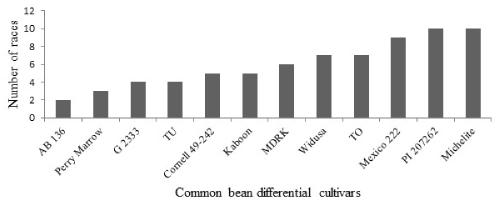
No single differential cultivar was resistant to all of the isolates collected and tested. Nevertheless, the differential cultivar AB 136 was a relatively more resistant cultivar than the rest, considering the number of races (only two, i.e. 1172 and 1250) that infected it. This observation is similar with findings of Sicard, et al. [22] and Gonzalez, et al. [23] who reported the differential cultivar AB 136, containing resistance two genes Co-6 and Co-8 as resistant to many isolates of C. lindemuthianum originating from different parts of the world. The differential cultivars Perry Marrow, G 2333 and TU were susceptible to three, four and four races, respectively, and ranked second and third most resistant cultivars to the pathogen races in the current experiment (Figure 3). A similar result was reported previously by Cláudia, et al. [24], where the cultivars AB 136 and G 2333 were found resistant to races collected from Paraná State of Brazil. Likewise, the differential cultivar TU was also reported to be susceptible to four isolates of C. lindemuthianum in another study [19].
On the other hand, the differential cultivars Michelite and PI 207262 were found to be the most susceptible differential cultivars showing susceptible reaction to 10 races, followed by Mexico 222, TO and Widus, which were susceptible to 9, 7 and 7 races in that order. More or less similar results were also reported by Cláudia, et al. [24] and Ansari, et al. [19]. The differential cultivars Michelite and Mexico 222 showed susceptible reaction to all of the 10 isolates collected from Grosso State, Brazil [25]. Also, according to Gonzaz (2016) [26], the bean differential cultivar Michelite was reported susceptible to all C. lindemuthianum isolates, which were collected from Arumeru, Karatu, Mbulu and Babati rural areas.
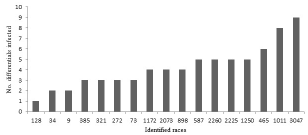
Of the 17 C. lindemuthianum races identified in the current work, race 3047 and race 1011 were the most 'cosmopolitan' races, infecting nine and eight differential cultivars, respectively, whereas races 128, 34 and 9 were 'narrow ranged', infecting a maximum of two cultivars, out of the total 12 cultivars (Figure 4).
With reference to bean gene pool, among the 17 identified races, 11 races (3047, 587, 2225, 1011, 2073, 465, 898, 1250, 2260, 1172 and 272) were pathogenic to the common bean differential cultivars having gene pool from both Mesoamerican and Andean origin. It is common to see races infecting differential cultivars having both gene pools as reported in Gonzaz [26]. Race 34 was pathogenic only to the differential cultivars from Andean gene pool, and races 385, 9, 321, 73 and 128 were pathogenic only to Mesoamerican gene pool. In line with this result, race 73, which infected only the Mesoamerican origin in this study, was also shown to be pathogenic to the differential cultivars from Mesoamerican origin [27,24]. Most of the isolates collected from Argentina and Central America were predominantly pathogenic to the Mesoamerican cultivars, while isolates collected from Africa, Europe, the Dominican Republic and South America showed a higher frequency of pathogenicity to the Andean cultivars than to the Mesoamerican differential cultivars [19].
Conclusion
The current study indicated that Ethiopian Colletotrichum lindemuthianum populations were highly variable as evidenced by the occurrence of 17 races of the pathogen from 30 isolates collected from five districts of south and central Ethiopia. Both highly virulent as well as less virulent races were observed. Current results also suggested that the Colletotrichum lindemuthianum population in Ethiopia could possibly be composed of highly virulent races that can cause much damage even to resistant/tolerant germplasms. The differential cultivar AB 136 was relatively the most resistant cultivar as it was susceptible only to two races. The differential cultivars Michelite and PI 207262 were the most susceptible differential cultivars as both showed susceptible reaction to 10 races. With reference to gene pool, race 34 was the only race that was pathogenic only on the differential cultivar from Andean gene pool, whereas races 385, 9, 321, 73 and 128 were pathogenic only on the differential cultivar from Mesoamerican gene pool. In conclusion, the current study suggested that the Ethiopian population of Colletotrichum lindemuthianum shall be regularly monitored for emergence of new races and race identification study need to be continued with more advanced techniques, i.e. molecular characterization method and by including more isolates across agro-ecological gradients of Ethiopia.
Acknowledgement
Authors thank the Ethiopian Institute of Agricultural Research (EIAR) for financial support. We also acknowledge Dr. Berhanu Amsalu for facilitation of acquisition of the differential cultivars from CIAT. We further extend our thanks to CIAT for the supply of initial seeds.
References
- Amin M (2013) An Overview of distribution, biology and the management of common bean anthracnose. J Plant Pathol Microb 4: 1-6.
- Amin M, Amare A, Nigusie D (2014) Effect of integrated management of bean anthracnose (Colletotrichum lindemuthianum) through soil solarization and fungicide application on epidemics of the disease and seed health in Hararghe highlands, Ethiopia. J Plant Pathol Microb 4: 182.
- Fitsum S, Amin M, Thangavel S, et al. (2014) Field management of anthracnose (Colletotrichum lindemuthianum) in common bean through foliar spray fungicides and seed treatment bioagents. Sci Technol Arts Res J 3: 19-25.
- Perseguini JMKC, Oblessuc PR, Rosa JRBF, et al. (2016) Genome-wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PLos One 11.
- Fernandez M, Casares A, Rodriguez R, et al. (2000) Bean germplasm evaluation for anthracnose resistance and characterization of agronomic traits: A new physiological strain of Colletotrichum lindemuthianum infecting Phaseolus vulgaris L. in Spain. Euphytica 114: 143-149.
- Batureine MJ (2009) Diversity of Colletotrichum lindemuthianum and reaction of common bean germplasm to anthracnose disease. Makerere University.
- Tesfaye B (1997) Less assessment study on haricot bean due to anthracnose. Pest Management Journal of Ethiopia 1: 69-72.
- Mahuku SG, Riascos JJ (2004) Virulence and molecular diversity within colletotrichum lindemuthianum isolates from Andean and Mesoamerican bean varieties and regions. European Journal of Plant Pathology 110: 253-263.
- CIAT (International Center for Tropical Agriculture) (1997) Bean program annual report 1995. Working document No. 163. Centro Internacional de Agriculture Tropical Colombia.
- Pastor-Corrales MA, Otoya MM, Molina A (1995) Resistance to Colletotrichum lindemuthianum isolates from Middle America and Andean South America in different common bean races. Plant disease 79: 63-67.
- Moses JK, Aston E, Vincent K, et al. (2016) Pathogenic variation of colletotrichum lindemuthianum causing anthracnose of beans (Phaseolus vulgaris) in Uganda. ESci Journal of Plant Pathology 5: 89-98.
- Bigirimana J, Hofte M (2001) Bean anthracnose: Inoculation methods and influence of plant stage on resistance of Phaseolus vulgaris cultivars. Journal of Phytopathology 149: 403-408.
- Allen DJ, Buruchara RA, Smithson JB (1998) Diseases of Common bean. The pathology of food and pasture legumes. CAB International 179-265.
- Pastor-Corrales MA (2004) Review of co-evolution studies between pathogens and their common bean hosts: Implication for the development of disease-resistant beans. Ann Rep Bean Improvement Coop 47: 67-68.
- Kelly JD, Vallejo VA (2004) A Comprehensive review of the major genes conditioning resistance to anthracnose in common bean. HortScience 39: 1196-1207.
- Habtu A, Sachet I, Zadoks JC (1996) A survey of cropping practices and foliar diseases of common beans in Ethiopia. Crop Protection 15: 179-186.
- Tesfaye B (2005) Colletotrichum lindemuthianum on beans in Ethiopia. Geographical distribution, importance, pathogenic variation, and management. In: PABRA Millennium Workshop 176-181.
- Genchev D, Christova P, Kiryakov I, et al. (2010) Breeding of common bean for resistance to the physiological races of anthracnose identified in Bulgaria. Biotechnology 24: 1814-1823.
- Ansari KI, Palacios N, Araya C, et al. (2004) Pathogenic and genetic variability among Colletotrichum lindemuthianum isolates of different geographic origins. Plant Pathology 53: 635-642.
- Gonçalves-Vidigal MC, Thomazella C, Elias HT, et al. (2008) Characterization of colletotrichum lindemuthianum isolates using differential cultivars of common bean in Santa Catarina state, Brazil. Braz arch biol Technol 51: 883-888.
- Mahuku GS, Jara CE, Cajiao C, et al. (2002) Sources of resistance to colletotrichum lindemuthianum in the secondary gene pool of phaseolus vulgaris and in crosses of primary and secondary gene pools. Plant dis 86: 1383-1387.
- Sicard D, Michalakis Y, Dron M, et al. (1997) Genetic diversity and pathogenic variation of Colletotrichum lindemuthianum in the three centers of diversity of its host Phaseolus vulgaris. Phytopathology 87: 807-813.
- Gonzalez M, Rodriguez R, Zavala ME, et al. (1998) Characterization of mexican isolates of colletotrichum lindemuthianum by using differential cultivars and molecular markers. Phytopathology 88: 292-299.
- Cláudia T, Maria Celeste GV, Pedro Soares, et al. (2002) Characterization of Colletotrichum lindemuthianum races in Paraná State, Brazil. Crop Breeding and Applied Biotechnology 2: 55-60.
- Gonçalves-Vîdigal MC, Nunes MPBP, Cruz AS, et al. (2010) Characterization of Colletotrichum lindemuthianum isolates from Matogrosso State, Brazil. Brazil 52-53.
- Gonzaz KK (2016) Identification of colletotrichum lindemuthianum and introgression of its resistance gene(s) to common bean (Phaseolus vulgaris L.) adapted in Tanzania. Sokoine University of Agriculture. Morogoro, Tanzania.
- Balardin RS (1997) Identification of physiological races of Colletotricum lindemutianumin Rio Grande, Brazil. Fitopathologia Brassilia 22: 50-53.
Corresponding Author
Abebe Gezahegn, Melkassa Agricultural Research Center, P.O. Box 436, Adama, Ethiopia.
Copyright
© 2021 Gezahegn A. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.