Morpho-Genetic Assessment of Greengram [Vigna radiata (L.) Wilczek] Landraces of Odisha, India for MYMV Resistance
Abstract
Green gram (Vigna radiata L. Wilczek) is seriously affected by Yellow Mosaic Disease (YMD) in tropical and subtropical countries of the world. The present investigation was carried out to screen the 52 indigenous mungbean germplasms and four breeding genotypes for the identification of Mungbean Yellow Mosaic Virus (MYMV) resistant. Based on twenty-four morpho-agronomic traits, it was observed that IPM-02-3 is highly resistance and five landraces i.e. Nadika local, Makarjhola local, Nayagarh local A, Gania local mixed, Jagasinghpur local-1 are moderately resistance to MYMV. There was a great variation among the landraces with regard to disease incidence and seed weight. Based on MYMV linked SSR markers, out of 56 genotypes including 52 landraces, 05 landraces (Nadika local, Jasinghpur local-1, Gania local mixed, Dhenkhanal 1Makarjhola local) are moderately resistant and one breeding genotype (IPM-02-14) act as resistant to MYMV. This study will helpful for conservation and management of genetic resource management of mungbean useful for breeding program.
Keywords
Mungbean, Landraces, SSR markers, MYMV, Molecular analysis
Introduction
Mungbean (Vigna radiata L. Wilczek) is widely cultivated throughout the Asia. It is the third most important pulse crop in India and covers more than 14% area under pulse cultivation and productivity rate of 461 kg/ha (NFSM pulses 2018-19) [1]. Mungbean seeds are rich source of minerals like calcium, iron, magnesium, phosphorous, potassium, vitamin A and Vitamin C. Being a 24% of dietary protein with low flatulence, and they are inseparable ingredients in the diets of the Asian population. The worldwide yield performance of mungbean is very low and its production has not considerably increased yet. The low yield is due to the susceptibility to insects and diseases caused by fungus, virus, or bacteria. Mungbean yellow mosaic virus (MYMV) (Begomo virus) is a major exotic disease of mungbean causing leaf discolouration and yield losses [2]. Developing MYMV-resistant mungbean varieties through classical breeding methods remain unsuccessful due to the rapid evolution of new isolates of MYMV and also the complexity mechanism MYMV resistance. Hence, screening based on natural occurrence in the hot spot areas is one of the viable methods and coupled with precision breeding using molecular markers. Genetic diversity among the germplasms, mining the markers linked for resistant gene and the QTL maps through molecular markers, has increased the efficiency in the breeding programs conferring resistance for MYMV [3,4]. Mungbean is drastically affected by the mungbean yellow mosaic virus and powdery mildew [5-7]. Another major disease reducing the crop productivity is the Cercospora leaf spot (CLS). To identify the genotype having a high level of resistant using molecular techniques has been reported [8-10]. Molecular markers are now widely used to identify loci and genome regions in many important crops including legume crops [11,12]. The present study is to identify the mungbean landraces resistant to MYMV by using agro-morphological traits and MYMV linked SSR markers.
Materials and Methods
Collection of genetic materials
A set of 52 mungbeans (Vigna radiata) landraces and four breeding genotypes were collected from the Centre for Pulses Research, Odisha University of Agriculture & Technology, Berhampur (Table 1). All the landraces were derived from different agro-climatic regions of Odisha, India (Figure 1) for genetic assessment study. Augmented design was adapted for the field experiment. The seeds were grown in the six rows of with 30 × 10 cm spacing with three replications. The experiment conducted in two consecutive summer seasons in the year 2017 and 2018. The rows distance was 30 cm apart and plant to plant distances were kept 10 cm. Two rows of the susceptible check are raised all around the experimental plot to attract whitefly and enhance infection of MYMV under field conditions. Insecticides and fungicides have not applied during the entire crop growth period. The weeding operation and other recommended packages and practices have been adapted.
Collection of phenotypic traits
Distinctness, uniformity and stability (DUS) guidelines [13] were adopted to collect the phenotypical traits after 30-d of sowing. The occurrence of whitefly and development of yellow Mosaic Disease (YMD) were regularly monitored in the crop. The disease infection and severity of MYMV was recorded and each plant rated on a 1 to 9 scale as per the guidelines presented in Table 2 [14]. The disease scoring recorded in two growth phases (vegetative and repro-ductive). Disease index has been calculated as per the formula is given below:
PDI = Sum of the numerical values/Total number of leaves examined × maximum grade value × 100
MYMV reaction (%) = Number of plant infected/Total number of plants × 100
Isolation and purification of genomic DNA
Young immature leaves were collected from field-grown plants and kept in -80 ℃ deep freezer till the extraction. Extraction of DNA was carried out as per the method of Doyle and Doyle [15]. Genomic DNA was isolated by using CTAB extraction buffer. The DNA pellet was washed with 70% (v/v) ethanol and dried properly. DNA pellet was properly dissolved with 250 μl of Tris-EDTA buffer (pH 8.0). To remove the RNA from the extracted sample, 5 μl of RNase (10 mg/ml) (Sigma, USA) was added in the DNA sample and incubated in the water bath at 37 ℃ for 1h. The DNA was further purified by extracting twice with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1). For precipitation of DNA, the one-tenth volume of 3M sodium acetate and an equal volume of absolute chilled ethanol were added for precipitation. The purified DNA pellet was collected and dissolved in 200 μl TE buffer at room temperature. DNA concentration was determined through 0.8% (w/v) agarose gel-electrophoresis stained with ethidium bromide by comparing the standard DNA (λ DNA digested by Hind-III) (Bangalore Genei, India).
PCR analysis
A gradient set up was made with a different annealing temperature of the synthesis primers. The range of annealing temperature and primer concentration can also further modified to get scorable amplification. The amplification was performed in a programmable gradient thermal cycler (Bio-Rad, USA) with an initial denaturation at 94 ℃ for 3 min followed by 35 cycles of denaturation at 94 ℃ for 1 min, annealing at a gradient temperature for 1 min, and extension at 72 ℃ for 2 min. The final extension was made for 10 min at 72 ℃. The gel electrophoresis was run with the amplified products in a 1.5% (w/v) agarose gel electrophoresis. The size of amplification was determined with the DNA ruler.
Statistical analysis
To ascertain the utility of observations based on data obtained from phenotypical and agronomic traits as per the DUS descriptors was analyzed. The resultant matrix was subjected to generate a dendrogram using the software program NTSYS pc Ver 2.2. Exeter Software, New York to estimate the genetic dissimilarity between genotypes [16].
Results and Discussion
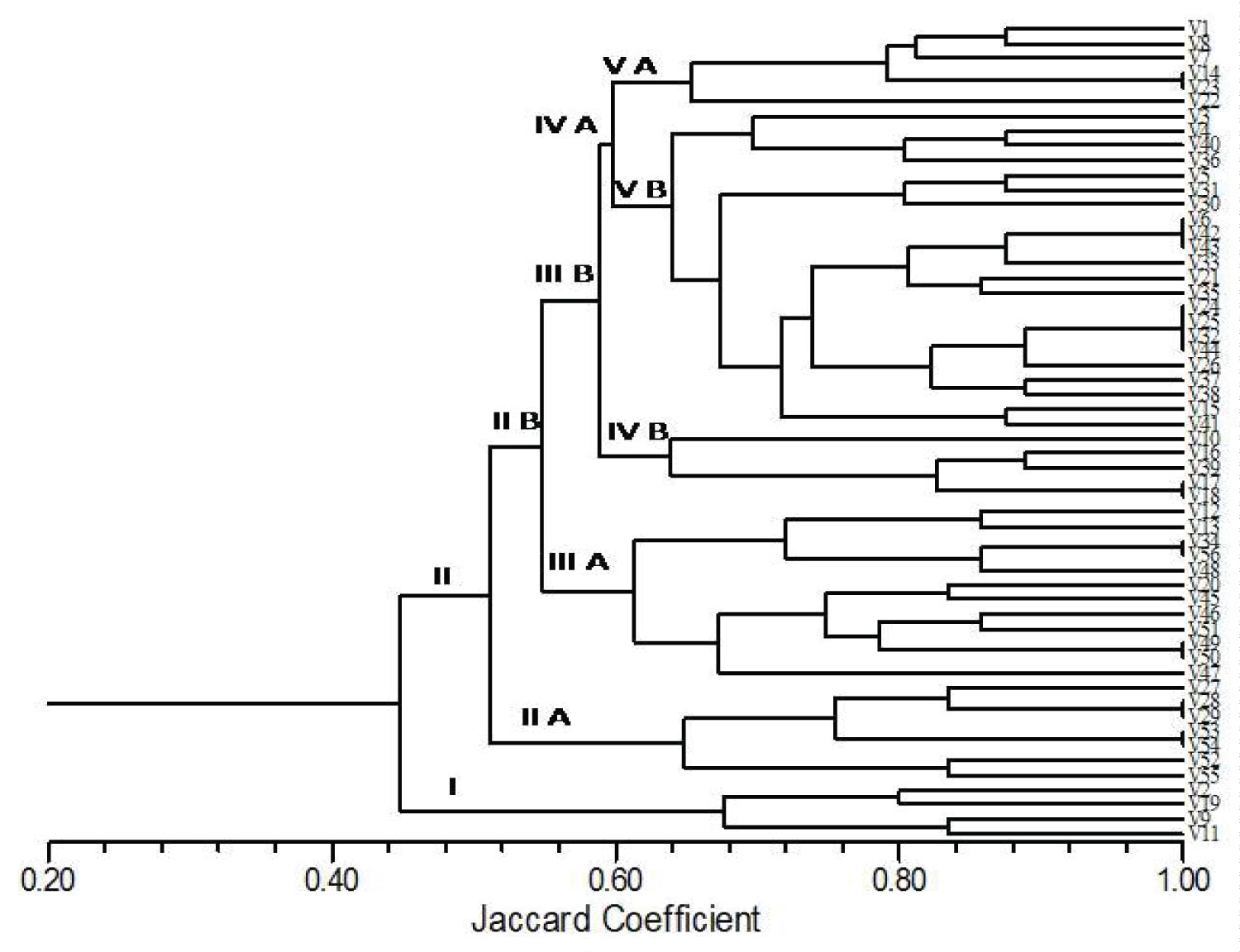
Fifty-two indigenous landraces and four breeding mungbean genotypes were evaluated under the natural condition in two conjugative years. The present results highlight the report of both morphological and agronomic traits of 56 genotypes including 52 indigenous landraces (Table 3). On the basis of the phenotypic characteristics such as plant height, growth habit, time of flowering, stem color, stem pubescence, leaf vein color, pod pubescence, pod position, pod length, seed luster and 100 seed weight and size of the seeds were greatly varied among the tested germplasms. The major phenotypic and agronomic traits such as seed color, seed lusture and seed weight and MYMV reaction were varied. It was observed that out of the tested germplasms screened, 34 are susceptible, 11 highly susceptible, 02 moderately susceptible, 02 resistant and 07 moderately resistant to MYMV (Table 4). The results implied that some of the indigenous landraces were higher yield (100g seed weight) and variation in degrees of MYMV reaction. These landraces include Sundargarh local-1 (4.3 g/100 seed weight), Nayagarh local green (3.7g), Nayagarh local A (3.1g), Bilipara local (3.4g), Banpur local green (3.1g), Saline 5 (3.0g.) and four breeding genotypes (Table 4). The lowest yield was observed in 'Jarsuguda local green' (2.3g/100 seed weight). On the basis of Disc Similarity Coefficient analysis, a cluster was made using 24 morpho-agronomic traits among 52 indigenous landraces and four breeding genotypes. It showed that there are two major clusters (I & II) among 56 tested germplasms (Figure 2). The first major cluster (I) having 52 genotypes with 44% similarity with cluster-II having four genotypes (V2, V9, V11, and V19). The second major cluster (II) further divided into two sub-major clusters i.e. II A & II B. Sub major cluster-IIA having only 07 genotypes and the second sub major cluster (II B) with45 genotypes with 50% similarity. Further, cluster-IIB divided into two minor clusters i.e, III A and III B. Cluster III A having 12 genotypes with 56% similarity with cluster III B (33 genotypes). Minor cluster-III B further divided into two sub minor clusters i.e. IV A and IV B. Cluster IVA consisted of five genotypes and cluster IVB having 28 genotypes with 58% similarity. Cluster IV B further divided into two clusters i.e. VA and VB. Cluster VA having 22 genotypes and VB with 10 genotypes having 64% similarity. The highest similarity (100%) was observed between V53 & V54, V28 & V29, V24 & V25, V42, V43 & V6 and V14 & V21 (Figure 2). The phenotypic and agronomic variation among the greengram genotypes has been reported [17,18].
Further, these landraces were assessed by using MYMV disease incidence on the basis of the rating scale. All genotypes were predicted from disease reaction with respect to R loci. The experiment was conducted both in Kharif and Rabi seasons for YMD incidence during the vegetative and reproductive phase. Gupta, et al. [19] reported that the appearance of MYMV incidence in mungbean under natural condition is dependent on the virus vector population and climatic conditions including temperature. The MYMV disease scoring was recorded as per the data 1 to 9 arbitrary scale. It was observed that MYMV infection was maximum visibility in the reproductive stage as compared to the vegetative stage. On the basis of disease scoring, the 52 indigenous landraces and four breeding genotypes were classified into five categories i.e. highly resistance (IPM-02-3), resistance (IPM-02-14 & OBGG-52), moderately resistance (Nadika local, Makarjhola local, Nayagarh local A, Gania local mixed, Jagasinghpur local 1), thirty-five were susceptible, two moderately susceptible (Nayagarh 4 , saline 5) and highly susceptible (Charpalli local, Bhapur local Black, Kantapada local brown, Purusottampur local, Berhampur local 2, Sundargarh local 1, Mahimunda local, Saline 1, Odogaon local Brown, Jharsuguda 1, Saline 7). There was positive correlation of the whitefly population in 20-30 days of crop growth and occurrence of disease incidence at 45-days-old crop with maximum temperatures [20]. Majority of indigenous landraces (about 62.5%) were included under susceptible, 3.5% moderately susceptible and 19.7% highly susceptible group.
Validation of germplasms through MYMV linked SSR markers
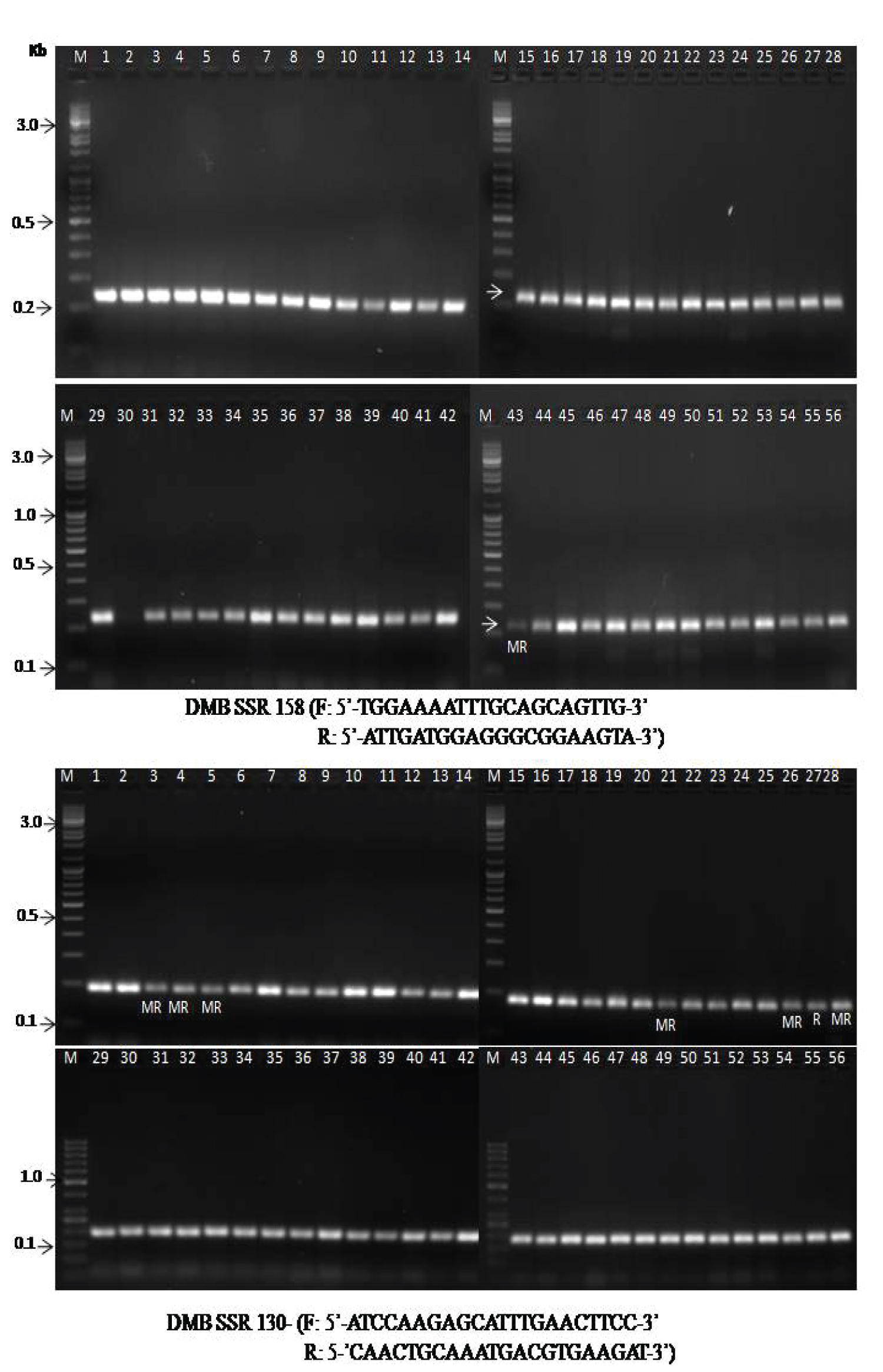
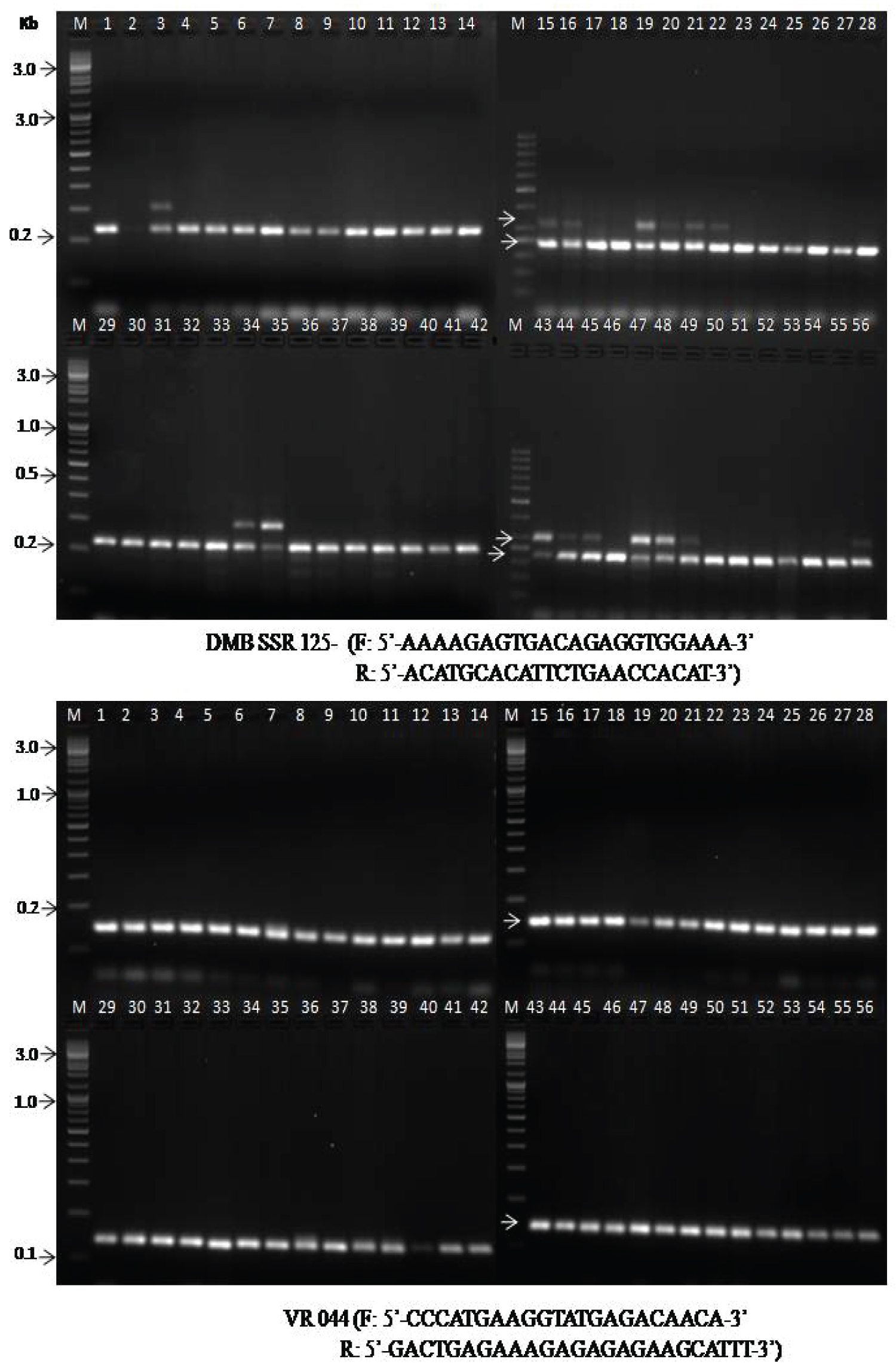
Validation of MYMV linked marker with the desired trait is an essential requirement for MAS in an advance breeding program [21,22]. They reported that the validation of the marker would help the identification of the resistant genotypes in the mapping population for breeding purposes. Various researchers have undergone research to know the molecular mechanism linked to MYMV for identification of germplasms and the movement of pathogens into the cell machinery for their survival. However, very scanty publications on molecular characterization of MYMV in mungbean genotypes [23-27]. The information related to genetic tools to clone and characterize R genes was not fully standardized in legume crops. About twenty selected markers have been used to identify the mungbean resistant to MYMV [19,28]. Out of 20 SSR markers, 16 markers amplified in the range of 120 to 280 bp which able to distinguish between resistant and susceptible genotypes (Table 5). Primer DMB-SSR-158 produced one amplicon at 220 bp in most of the landraces including breeding genotypes. Based on the amplification, it was observed the landrace i.e. Nadica local was moderately resistant to MYMV. Primer DMB-SSR-130 produced one amplicon at 190 bp in most of the selected landraces. Based on the amplification, it was observed that four landraces (Jasinghpur local-1, Gania local mixed, Dhenkhanal 1 and Makarjhola local) were moderately resistant and check genotype IPM-02-14 as resistant to MYMV (Figure 3A). The primer DMB-SSR-125 generated two amplicons at 220 bp and 240 bp. However, 240 bp amplicon indicates in 12 landraces. Primer VR-044 and VR-078 showed one amplicon each at 150 bp and 130 bp respectively in all the landraces and breeding genotypes. Kang, et al. [29] reported that about 80% of viral resistance is monogenically controlled by the host's resistant factors. In most of the cases, the R genes derived from monogenic dominant resistance plants successfully linked to molecular markers from consensus motifs of other resistance (R) gene or R gene homologous sequences as reported earlier [21,30,31]. The primers like DMB-SSR-130, DMB-SSR-158, DMB-SSR-125, VR-044, and VR-078 were completely linked to MYMV resistance and shows amplicon at 190 bp, 220 bp, 240 bp, 150 bp, and 130 bp respectively (Figure 3B).
Conclusions
It is concluded that the screening of indigenous landraces including breeding genotypes had significant differences with regard to yield attributing traits and phenotypic characteristics. Based on the morpho-agronomic screening of 56 genotypes including 52 landraces, IPM-02-14, OBGG-52 was resistant, IPM-02-3 highly resistance, and five moderately resistance (Nadika local, Makarjhola local, Nayagarh local A, Gania local mixed, Jagasinghpur local 1) to MYMV. On the basis of MYMV linked SSR markers, five mungbean landraces (Nadica local, Jasinghpur local 1, Gania local mixed, Dhenkhanal 1 and Makarjhola local) were moderately resistant and breeding genotype IPM-02-14 as resistant to MYMV. The present investigation revealed that both morpho-agronomic screening and SSR based linked markers are efficient for the identification of MYMV-resistant genotype which will helpful for an advanced breeding program in mungbean. Identified landraces shall be considered further as a resistant donor for MAS.
Acknowledgements
The authors wish to acknowledge to Department of Biotechnology, Ministry of Science and Technology, Govt. of India, New Delhi for PG-HRD teaching program and Department of Science & Technology for proving the FIST grant for development of infrastructure.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- FAO (2019) FAO statistics.
- Fauquest CM, Bridden RW, Brown JK, et al. (2008) Geminivirus strain demarcation and nomenclature. Arch Virol 153: 783-821.
- Sudha M, Karthikeyan A, Nagarajan P, et al. (2013) Screening of mungbean (Vigna radiata) germplasms for resistance to mungbean yellow mosaic virus using agroinoculation. Can Jour Plant Pathol 46: 717-723.
- Grover DK, Weinberger K, Shanmugasundaram S, et al. (2005) Constraint analysis of mungbean production incereal-based cropping system in Punjab. Jour Res Punjab Agriculture University 42: 223-231.
- Chankaew S, Somta P, Isemura T, et al. (2013) Quantitative trait locus mapping reveals conservation of major and minor loci for powdery mildew resistance in four sources of resistance in mungbean [Vigna radiata (L.) Wilczek]. Mol Breeding 32: 121-130.
- Gupta SK (2008) Pulse production in India- Constraints and Opportunities. In: Glimpses of Indian Agriculture: Macro and Micro Aspects. Academic Foundation 1: 206-250.
- Varma A, Malathi VG (2003) Emerging germinivirus problems: A serious threat to crop production. Ann App Biol 142: 145-164.
- Selvi R, Muthiah AR, Manivannan N, et al. (2006) Tagging of RAPD marker for MYMV resistance in mungbean (Vigna radiata (L.) Wilczek. Asian J of Plant Sci 5: 277-280.
- Taggar HGK, Gill RS, Sandhu JS (2013) Evaluation of blackgram [Vigna mungo (L.) Hepper] genotypes against whitefly, B. tabaci (Gennadius) under greenhouse conditions. Acta Phytopathologica et Entomologica Hungarica 48: 53-62.
- Sulistyo A, Inayati A (2016) Mechanisms of antixenosis, antibiosis and tolerance of fourteen soyabean genotypes in response to whiteflies (Bemisia tabaci). Biodiversity 17: 447-453.
- Kumar SV, Tan SG, Qush SC, et al. (2002) Isolation of microscatellite in mungbean, Vigna radiata . Mol Eco Notes 2: 96-98.
- Kumar SV, Tan SG, Qush SC, et al. (2002) Isolation of microsatellite markers in mungbean, Vigna radiata. Mol Eco Notes 2: 293-295.
- PPV & FRA (2007) Protection of plant varieties and farmer's rights Authority, Guidelines for the conduct of test for the Distinctness, Uniformity and Stability on Blackgram (Vigna mungo L.) Hepper, Plant Variety Journal of India 1: 4-6.
- Singh G, Sharma YR, Kaur L (1992) Method of rating mungbean yellow mosaic virus of mungbean and urdbean. Plant Dis Res 7: 1-6.
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bulletin 19: 11-15.
- Rohlf FJ (2005) NTSYSpc (Numerical Taxonomy & Multivariate Analysis System). Version 2.2, Exeter Software, Applied Biostatistics Inc., New York.
- Abna F, Golam F, Bhassu S (2012) Estimation of genetic diversity of mungbean (Vigna radiata (L.) Wilczek) in Malaysian tropical environment. Afr Jour Microbiol 6: 1770-1775.
- Gadakh SS, Dethe AM, Kathale MN, et al. (2013) Genetic diversity for yield and its component traits in green gram [Vigna radiata (L.) Wilczek]. Journal of Crop and Weed 9: 106-109.
- Gupta S, Gupta DS, Anjum TK, Pratap A, et al. (2013) Inheritance and molecular tagging of MYMV resistance gene in black gram (Vigna mungo L. Hepper). Euphytica 193: 27-37.
- Murugesan S, Chelliah S (1977) Influence of sowing time on the incidence of the vector Bemisia tabaci (Genn.) and the yellow mosaic disease of greengram. Madras Agric Jour 64: 128-130.
- Maiti S, Basak J, Kundagrami S, et al. (2011) Molecular marker-assisted genotyping of mungbean yellow mosaic India virus resistant germplasms of mungbean and urdbean. Mol Biotechnol 47: 95-104.
- Binyamin R, Khan MA, Khan AI, et al. (2016) Genetic diversity of mungbean genotypes in relation to resistance against mungbean yellow mosaic virus. Pak J of Botany 48: 1273-1277.
- Gupta OM (2003) Resistance to Mungbean yellow mosaic virus, phenotypic characters and yield components in Urdbean. Indian Phytopathol 56: 110-111.
- Shivaprasad P, Thillaichidambaram P, Balaji V, et al. (2006) Expression of full length and truncated Rep genes from mungbean yellow mosaic virus-Vigna inhibits viral replication in transgenic tobacco. Virus Genes 33: 365-374.
- Gupta S, Kumar S, Singh RA, et al. (2005) Identification of a single dominant gene for resistance to mungbran yellow mosaic virus in blackgram. SABRAO Jour Breeding and Genetics 37: 85-89.
- Gupta SK, Souframanien J, Gopalakrishna T (2008) Construction of a genetic linkage map of black gram based on molecular markers and comparative studies. Genome 51: 628-637.
- Maheshwari R, Panigrahi G, Angappan J (2014) Molecular characterization of distinct YMV (yellow mosaic virus) isolates affecting pulses in India with the aid of coat protein gene as a marker for identification. Mol Breeding Reports 41: 2635-2644.
- Prasanthi L, Reddy BVB, Gettha B, et al. (2013) Molecular markers for screening yellow mosaic disease resistance in black gram [Vigna mungo (L.) Hepper]. Electron Jour Plant Breeding 4: 1137-1141.
- Kang BC, Yeam I, Jahn MM (2005) Genetics of plant virus resistance. Annual Rev Phytopathol 43: 581-621.
- Diaz-pendon JA, Truniger V, Nieto C, et al. (2004) Advances in understanding recessive resistance to plant virus. Mol Plant Patheol 5: 223-233.
- Maule AJ, Caranta C, Boulton MI (2007) Sources of natural resistance to plant viruses:status and prospects. Mol Plant Pathol 8: 223-231.
Corresponding Author
Gyana Ranjan Rout, Department of Agricultural Biotechnology, College of Agriculture, Odisha University of Agriculture & Technology, Bhubaneswar-751003, Odisha, India.
Copyright
© 2021 Kumari GP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.