Influence of Acetaldehyde on Oral Epithelial Cells
Abstract
Acetaldehyde, a product of alcohol metabolism, has been recognized a risk factor for carcinogenesis. To clarify the carcinogenic effects of acetaldehyde in the oral cavity and mechanisms involved, normal oral epithelial cells were incubated with several concentrations of acetaldehyde and assays of genetic and epigenetic changes were performed. Acetaldehyde induced malignant transformation of normal oral epithelial cells and demethylation of the erbB2 gene. In addition, regulation of DNA methylation was suppressed when cells were subjected to alternating exposure to medium containing acetaldehyde and medium without acetaldehyde. These findings suggest that habitual alcohol drinking increases the risk of developing oral cancer, and that an appropriate abstinence period may reduce this risk.
Keywords
Oral Epithelial Cell, Acetaldehyde, Carcinogenesis, Epigenetics
Introduction
Cancer is one of the most common diseases worldwide, with a high annual mortality. Cancer is characterized by abnormal cell proliferation, gene mutation, metastasis to other organs, and invasion to mesenchymal tissue. Despite extensive research on the mechanisms involved, including carcinogenesis itself, many details remain unclear. Oral squamous cell carcinoma (OSCC) is an invasive epithelial neoplasm of the oral cavity showing various degrees of squamous differentiation. Kusama, et al. have previously shown that OSCC exhibits p53 mutation, and have suggested that this is involved in the early stage of the dysplasia-carcinoma sequence in oral squamous epithelium [1].
Epidemiological studies have shown that alcohol is a risk factors for carcinogenesis along with smoking [2,3]. When alcohol is ingested, it is metabolized to acetaldehyde by alcohol dehydrogenase (ADH), and then further to acetic acid and water by aldehyde dehydrogenase (ALDH) [4]. It is thought that the rate of DNA damage, and thus carcinogenesis, increases under circumstances where ALDH deficiency causes tissues to be exposed to acetaldehyde for a relatively long time. In addition, it has been reported that alcohol induces epigenetic change in oral epithelial cells, and that the acetaldehyde concentration in the oral cavity is much higher in drinkers than in non-drinkers, since certain oral bacteria produce acetaldehyde through alcohol decomposition [5,6]. These findings suggest that alcohol/acetaldehyde play a causative role in oral cancer.
The aim of the present study was to clarify the carcinogenetic effect of acetaldehyde in the oral cavity and the mechanisms involved.
Materials & Methods
Cell line
Normal Human Keratinocytes were purchased from Cell Research and incubated with RPMI1640 medium in a humidified atmosphere of 5% CO2 at 37 °C. For studies, cells were incubated with 0.01, 0.05, 0.1% of acetaldehyde for several days. The concentration of acetaldehyde was adjusted based on the blood equivalent percentage.
Malignant transformation assay
To study the effect of acetaldehyde to oral cells, malignant transformation assay was carried out by using CytoSelect 96-well cell Transformation Assay Kit (Cell Biolabs) in accordance with the protocol, and absorbance of 485/520 nm was measured following solubilized the agar.
DNA methylation assay
For DNA methylation assay, Methyl Flash Global DNA Methylation (5-mC) ELISA Easy Kit (EPIGENETEK) was used in accordance with the protocol. DNA which was isolated after the incubation was binded to assay wells and complexed with 5-mC Detection Complex Solution, then measured the absorbance of 450 nm.
Real-time RT-PCR
Real-time RT-PCR was performed by using a One-Step SYBR Prime Script RT-PCR Kit II (Takara), and Thermal Cycler Dice Real Time System (Takara) in accordance with the standard protocol. The primers, based on sequences for erbB, Myc, APC and p53, are shown at Table 1.
Bisulfate-PCR
DNA was treated with hydrogen sulfite bisulfate to base substitute cytosine for uracil. If the cytosine was methylated, base substitution did not occur because it was protected by the methyl group. PCR and electrophoresis were then performed using the specific primer for upstream of each gene (Table 2) and 2% agar gel.
Results
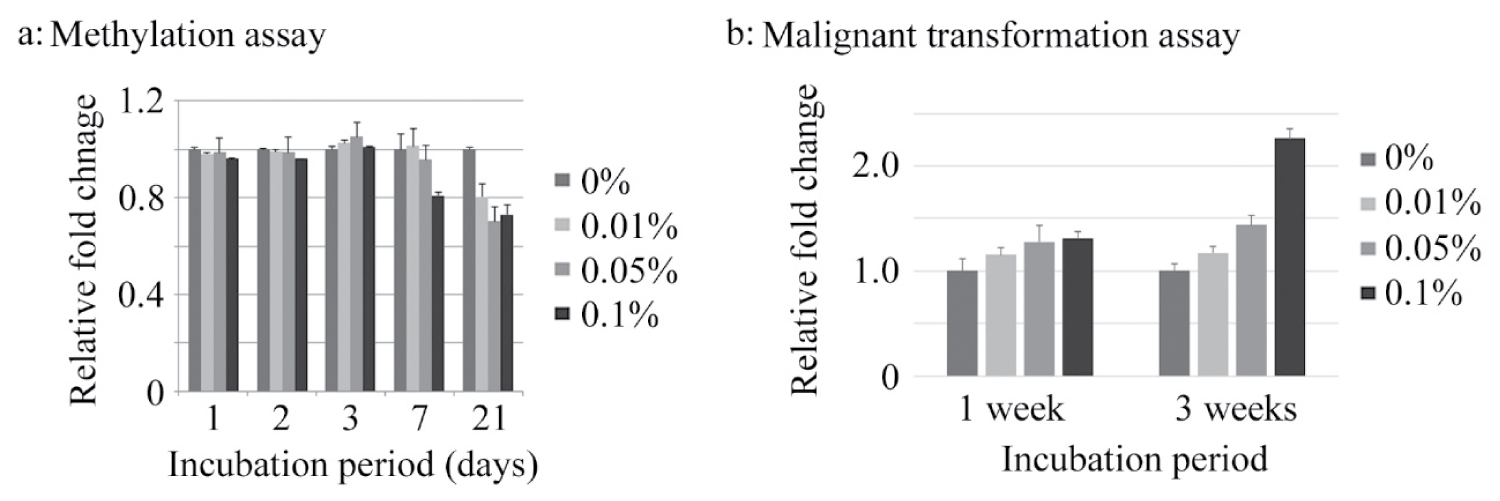
After incubation of cells in the presence of several concentrations of acetaldehyde for several days, DNA methylation assay was performed. Acetaldehyde tended to decrease DNA methylation in a dose- and time-dependent manner, especially 1 and 3 weeks incubation (Figure 1a). Then, malignant transformation assay was performed after 1 and 3 weeks incubation with acetaldehyde. The degree of malignant transformation tended to increase dose- and time-dependently (Figure 1b).
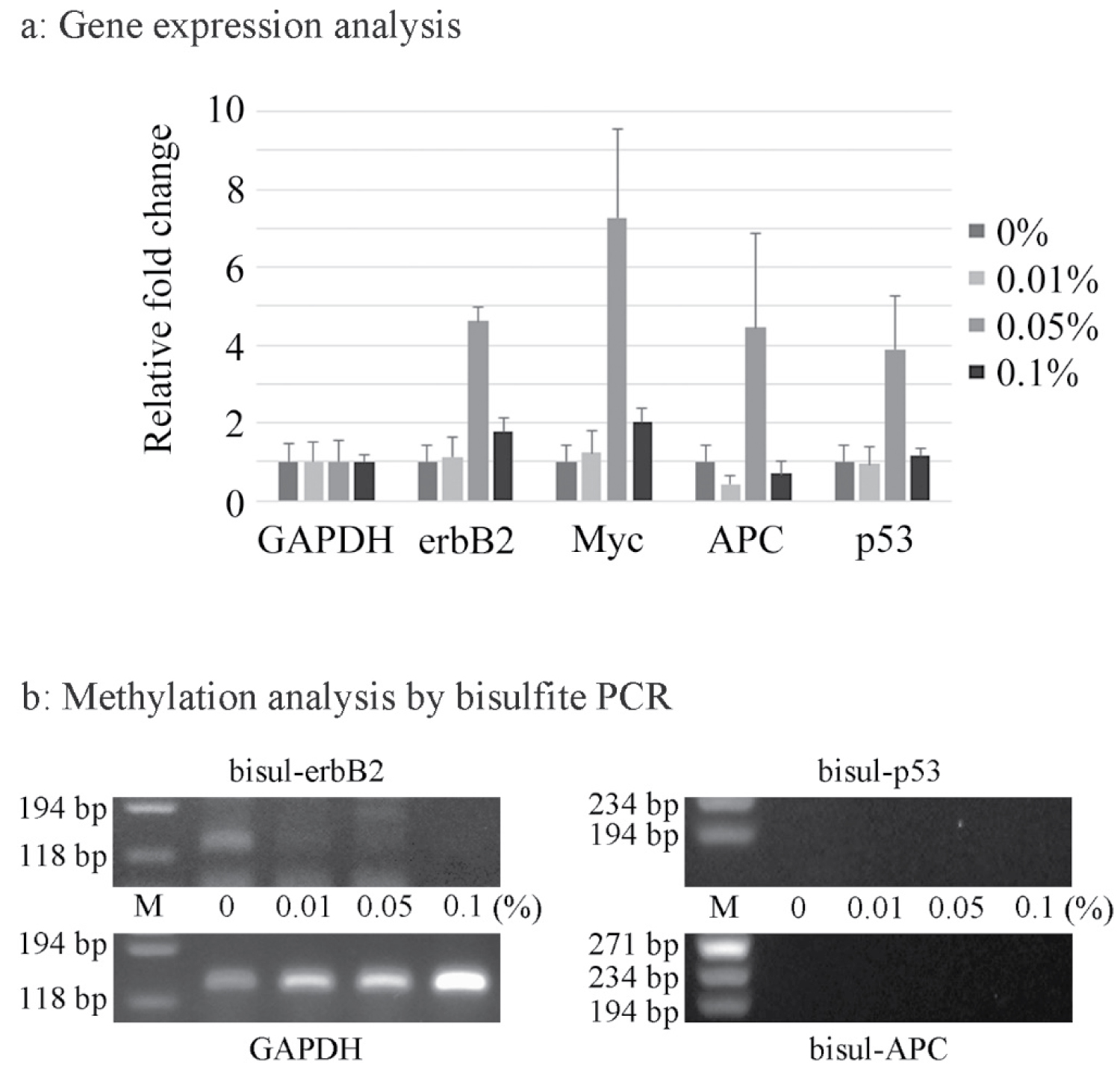
Gene expression analysis of erbB2, Myc, APC and p53 was performed by the real-time RT-PCR method. The expression levels of all genes examined were increased after exposure to 0.05% acetaldehyde, and the levels of erbB2 and Myc expression were slightly increased even after exposure to 0.1% acetaldehyde (Figure 2a). Bisulfate-PCR was then performed for evaluation of the methylation status. Although the APC and p53 genes were not detected, it was suggested that acetaldehyde induced demethylation of erbB2 (Figure 2b).
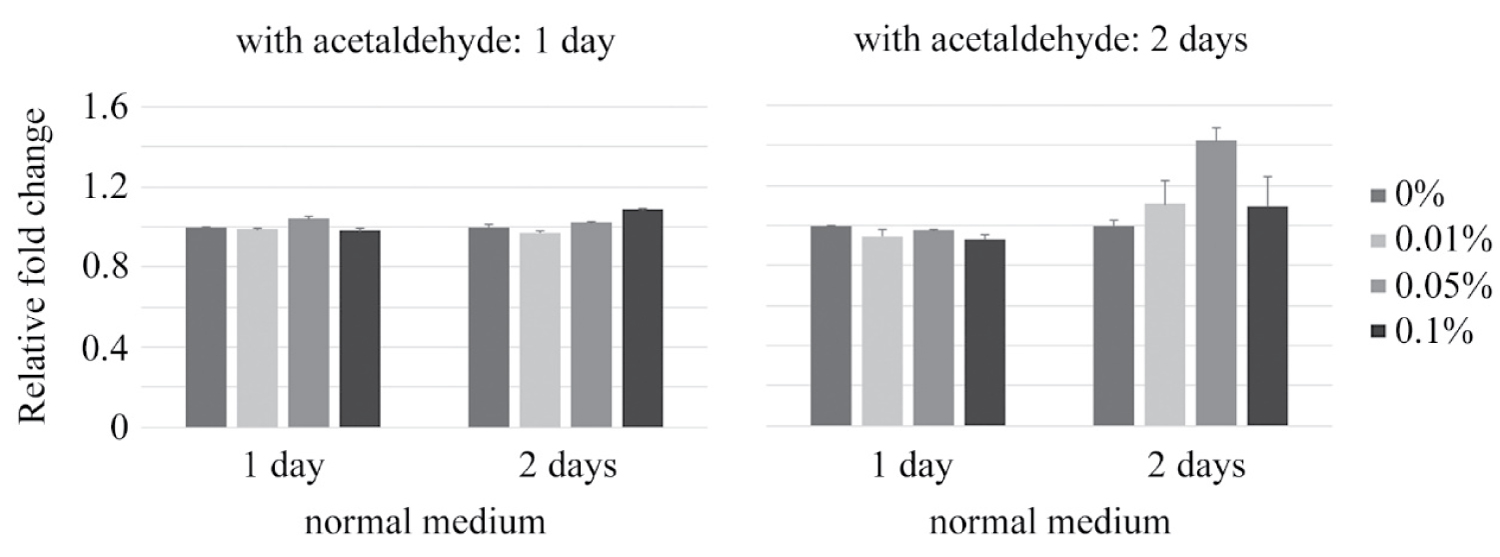
Medium containing acetaldehyde was exchanged for normal medium alternately every 1 or 2 days, and then DNA methylation assay was performed. The ratio of DNA methylation was up-regulated when medium exchange was conducted every 2 days (Figure 3).
Discussion
The oral cavity is an area frequently exposed to a variety of stimuli, including hard (sharp) sensation, heat, cold, and spiciness, and it is known that chronic mechanical stimulation can increase the possibility of carcinogenesis. The oral cavity is also one of the early routes of bacterial and/or viral infection, so it is easy to speculate that these foreign bodies may also contribute to carcinogenesis [7-10]. Thus, it is recognized that the oral cavity represents an area where a variety of carcinogens are constantly present, making it difficult to reveal the causes of oral carcinogenesis.
Although genetic mutations were not detected in present study, it has been reported that oral cancer carries mutations in the p53, PI3K and PTEN genes, and also in mitochondrial DNA [11-17], suggesting that acetaldehyde could be responsible for such mutations. Comprehensive gene expression analysis will be one of the challenges for the future.
In this study, the expression levels of the four genes examined were reduced in the presence of 0.1% acetaldehyde, perhaps due to the cytotoxic effect of acetaldehyde. Many other substances are known to exert such a cytotoxic effect, and normal cells are more susceptible to such effects than tumor cells in general. Although cells derived from oral cancer were not used in this study, acetaldehyde at concentrations higher than 0.1% may be needed in order to obtain meaningful experimental data for oral cancer cells.
Although a number of studies have investigated the Epigenetics of the EGFR gene in oral cancer [18-20], there is little information for other members of erbB gene family. In this study, expression and demethylation of the erbB2 gene were promoted by acetaldehyde. ErbB2 is a receptor type tyrosine kinase thought to form heterodimers with other erbB molecules to potentiate intracellular signals including the PI3K/AKT and RAS/MAPK pathways [21-24]. It is likely that promotion of the demethylation and expression of the erbB2 gene by acetaldehyde would lead to further proliferation of cells through the formation of more heterodimers. In addition, if genetic mutations occur in erbB2, the cell cycle would likely become perturbed, leading to progression of canceration.
It was suggested that methylation was promoted when oral normal cells were subjected to alternating culture in medium with and without acetaldehyde every two days. Contrary to demethylation, such promotion of methylation would be expected to suppress excessive gene expression, suggesting that moderate intervals of alcohol consumption with a rest day might have an inhibitory effect on excessive or extra gene expression.
Excessive alcohol consumption can contribute to a variety of diseases, including cancer, as well as creating social problems and disrupting relationships. However, there is an adage in Japan that says "alcohol is the best of a hundred medicines," and when consumed in moderation and at moderate intervals, it may help to reduce stress, prevent cancer, and build more amicable relationships.
Acknowledgement
This experiment was supported by a Miyata Research Grant from Meikai University.
Conflict of Interest
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Kusama K, Okutsu S, Takeda A, et al. (1996) p53 gene alterations and p53 protein in oral epithelial dysplasia and squamous cell carcinoma. J Pathol 178: 415-421.
- Blot WJ, McLaughlin JK, Winn DM, et al. (1988) Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res 48: 3282-3287.
- Baan R, Straif K, Grosse Y, et al. (2007) WHO International agency for research on cancer monograph working group: Carcinogenicity of alcoholic beverages. Lancet Oncol 8: 292-293.
- Matsuda T, Yabushita H, Kanaly RA, et al. (2006) Increased dna damage in aldh2-deficient alcoholics. Chem Res Toxicol 19: 1374-1378.
- Homann N, Jousimies-Somer H, Jokelainen K, et al. (1997) High acetaldehyde levels in saliva after ethanol consumption: methodological aspects abd pathogenetic implications. Carcinogenesis 18: 1739-1743.
- Vakevainen S, Tillonen J, Agarwal DP, et al. (2000) High salivary acetaldehyde after a moderate dose of alcohol in ALDH2-deficient subjects: Strong evidence for the local carcinogenic action of acetaldehyde. Alcohol Clin Exp Res 24: 873-877.
- Sand L, Jalouli J (2014) Viruses and oral cancer. is there a link? Microbes Infect 16: 371-378.
- Hubbers CU, Akgul B (2015) HPV and cancer of the oral cavity. Virulence 6: 244-248.
- Kikuchi K, Noguchi Y, Garcia-Nino de Rivera MW, et al. (2016) Detection of epstein-barr virus genome and latent infection gene expression in normal epithelia, epithelial dysplasia, and squamous cell carcinoma of the oral cavity. Tumour Biol 37: 3389-3404.
- Shillitoe EJ (2018) The microbiome of oral cancer. Crit Rev Oncog 23: 153-160.
- Rowley H, Sherrington P, Helliwell TR, et al. (1998) p53 expression and p53 gene mutation in oral cancer and dysplasia. Otolaryngol Head Neck Surg 118: 115-123.
- Liao PH, Lee TL, Yang LC, et al. (2001) Adenomatous polyposis coli gene mutation and decreased wildtype p53 protein expression in oral submucous fibrosis: A preliminary investigation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 92: 202-207.
- Mewara A, Gadbali AR, Patil S (2010) C-deletion mutation of the p53 gene at exon 4 of codon 63 in the saliva of oral squamous cell carcinoma in central india: A preliminary study. J Investig Clin Dent 1: 108-113.
- Ara N, Atique M, Ahmed S, et al. (2014) Frequency of p53 gene mutation and protein expression in oral squamous cell carcinoma. J Coll Physicians Surg Pak 24: 749-753.
- Walls M, Baxi SM, Mehta PP, et al. (2014) Targeting small cell lung cancer harboring PIK3CA mutation with a selective oral PI3K inhibitor PF-4989216. Clin Cancer Res 20: 631-643.
- Yuan RT, Sun Y, Bu LX, et al. (2015) Gene mutations in the d-loop region of mitochondrial DNA in oral squamous cell carcinoma. Mol Med Rep 11: 4496-5000.
- Shah S, Jajal D, Mishra G, et al. (2017) Genetic profile of PTEN gene in indian oral squamous cell carcinoma primary tumors. J Oral Pathol Med 46: 106-111.
- Rautava J, Jee KJ, Miettinen PJ, et al. (2008) ERBB receptors in developing, dysplastic and malignant oral epithelia. Oral Oncol 44: 227-235.
- Molinolo AA, Amornphimoltham P, Squarize CH, et al. (2009) Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol 45: 324-334.
- Dumache R, Rogobete AF, Andreescu N, et al. (2015) Genetic and Epigenetic Biomarkers of Molecular Alterations in Oral Carcinogenesis. Clin Lab 61: 1373-1381.
- Troiani T, Martinelli E, Capasso A, et al. (2012) Targeting EGFR in pancreatic cancer treatment. Curr Drug Targets 13: 802-810.
- Weigelt B, Lo AT, Park CC, et al. (2010) HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat 122: 35-43.
- Moasser MM (2007) The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26: 6469-6487.
- Wang Z (2017) ErbB Receptors and Cancer. Methods mol biol 652: 3-35.
Corresponding Author
Yuji Miyazaki, Division of Basic Biology, Department of Oral Biology and Tissue Engineering, Meikai University School of Dentistry, Japan 1-1 Keyaki-dai, Sakado, Saitama, 350-0283, Japan, Tel: +81-49-279-5101.
Copyright
© 2021 Miyazaki Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.