A Retrospective Study of Uropathogenic ESBL-producing Enterobacteriaceae among Hospitalized Patients, Khartoum-Sudan
Abstract
Uropathogenic extended-spectrum beta-lactamases-producing Enterobacteriaceae (ESBL-PE) remains a global cause of urinary tract infections (UTIs) among hospitalized patients as these enzymes hydrolysing 3rd generation cephalosporin's (3rd GC). Those bacteria may not respond to therapy and thus further limiting antimicrobial selections. This study aimed to detect uropathogenic Enterobacteriaceae producing ESBL genes and their antimicrobial resistance pattern. Hundred and four (n=104) Enterobacteriaceae uropathogens were isolated in (2007-2008), from midstream urine (MSU) samples. In 2019, 54% of the recovered isolates were resistant to 3rd GCs. The presence of ESBL-genes (blaTEM, blaSHV, and blaCTX –M genes) screened by multiplex polymerase chain reaction (PCR). ESBL gene producing isolates were detected among 44.6%, and 36% of the ESBL-producing isolates were harbouring 2 ESBL genes. The common gene was the blaTEM, 40%, followed by 16% blaCTX-M and 8% blaSHV. The most dominant isolate was E. coli 60%, followed by K. pneumonia 36%, and E. aerogenes 4%. This study revealed that blaTEM was the most prevalent ESBL-PE uropathogens before 10 years in our region, although the current ESBL genes is CTX-M types exceeded SHV and TEM as the dominating type, and is thought to involve clonal spread. Our community; needs molecular-based epidemiological studies to report the antimicrobial resistance genes in the Sudan region besides the origin of those genes.
Keywords
Extended-spectrum, ß-lactamases producing Enterobacteriaceae, uropathogens, bla-genes
Abbreviations
MSU: Mid-stream urine; GNB: Gram-negative bacteria; ESBL: Extended-spectrum β-lactamase; ESBL-PE: Extended-spectrum β-lactamase producing Enterobacteriaceae; 3rd GC: 3rd generation cephalosporin's; NCCLS: National committee on clinical laboratory standards; CLSI: Clinical laboratory standard institute; UTI: Urinary tract infection; MHA: Mueller Hinton agar; MDR: Multidrug resistance; Escherichia coli: E.coli; Klebsiella pneumoniae: K. pnuemoniae; Enterobacter aerogenes: E. aerogenes; Species: spp
Introduction
Enterobacteriaceae member commonly related to hospital acquired UTI infections, mainly catheter-related UTIs. They are caused particularly by Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates with high recurrence rates of increasing antimicrobial resistance among those uropathogens [1,2]. With the wide use of extended-spectrum cephalosporin's antibiotic in the healthcare setting, used as first-line therapy for Enterobacteriaceae UTI, the major worry is the increased reports on the dissemination of Hospital-Acquired UTI with ESBL-producing Enterobacteriaceae (ESBL-PE) that may limit available treatment options, as they contribute significantly to the rapid dissemination of resistant organisms and their genes [3,4]. The ESBLs are a β- lactamase with hydrolytic activity against penicillin's, extended-spectrum cephalosporins, and monobactams, and are inhibited by clavulanic acid. The predominant ESBL genotypes before 2000 were TEM and SHV and produced by Klebsiella spp., Enterobacter spp., and E. coli. Regarding the nature of ESBL dissemination has changed during the past few years, thought Enterobacteriaceae strains express CTX-M as the most common type of ESBLs mainly among E. coli strains [5,6]. They disseminate in isolates from the same species, as well as across different species. The localization of these genes on transmissible elements via horizontal gene transfer, especially the emergence of ESBL-PE, is of great concern due to their limited treatment options. While the spread of ESBL-PE starts to increase in our region due to extensive self-treatment as well as less effective infection control [7], whereas, still there are few available reports on hospital environments. Therefore, the present study aimed to detect the ESBLs genotype producing Enterobacteriaceae and the antimicrobial resistance patterns of clinical isolates associated with UTI among inpatients admitted to the Academy Charity Teaching Hospital (ACTH) in Khartoum State, Sudan. Such data make information available and serve to understand the ESBLs genes distribution in 2008.
Methods
Bacterial isolates
A retrospective study of 56 (54 %) of 3rd GCs resistant Enterobacteriaceae uropathogens isolates was recovered from preserved samples of inpatients with UTI from December 2007 to July 2008, admitted in the urology department of ACTH. An analytical profile index (API 20E-bioMerieux, Marcy-I'Etoile, France) was performed to identify Enterobacteriaceae member to species level for all Enterobacteriaceae uropathogens isolates. Enterobacteriaceae uropathogens counted ≥ 105 colony forming units (CFU)/mL of urine specimens were considered as a significant bacteriuria [8]. The isolates at that time preserved at -80℃. In March-May 2019; for recovery of the 25of phenotypically screened ESBL-producing Enterobacteriaceae uropathogens isolates; the Tryptose Soy Broth (HiMEDIA Laboratories Pvt. Ltd., Mumbai, India) used for re-suspension of the stored isolate, and the isolates were inoculated on MacConkey agar (HiMEDIA Laboratories Pvt. Ltd., Mumbai, India) and incubated for 18-24 h at 37℃.
Antimicrobial susceptibility test
Kirby Bauer's technique was done for antimicrobial susceptibility testing of ESBL-PE isolates and interpreted according to the standards from the CLSI (Clinical and Laboratory Standards Institute) [9]. The following antimicrobial agents have tested: cefotaxime (CTX: 30 μg), ceftazidime (CAZ: 30 μg), cefpodoxime (CPD: 10 μg), aztreonam (ATM: 30 μg) as ESBL screening tests. The following antimicrobial agents have tested amikacin (AK: 30 μg), ciprofloxacin (CIP: 25 μg), co-trimoxazole (COT:1.25/23.75 μg), gentamycin (CN: 10 μg) and nitrofurantoin (NIT: 300μg) (Oxoid, UK and Hi Media Laboratories Pvt. Ltd., India).
Genomic DNA extraction
The chromosomal DNA was extracted from ESBL-PE bacterial cells by guanidine chloride method [10]; and the concentration and purity of the extracted DNA were determined using NanoDrop-Spectrophotometer (Thermo-scientific, USA) at Central Laboratory, Ministry of High Education and Scientific Research. The chromosomal DNA was Stored at -20℃ for further use.
PCR amplification of ESBL genes
The presence of the ESBL genes (blaSHV, blaTEM and blaCTX-M), determined by PCR amplification for those encoding genes by using previously described primers (Table 1). For PCR protocol; 0.5 μl of each the forward and reverse primers (ESBL genes), 5 μl of the bacterial DNA, 8 μl water of injection was added to the 4 μl PCR Master-Mix (5X HOT FIREPol ®Blend Master Mix Ready to Load, Estonia) with a final volume of 20 μl. The reaction mixture as follows: Initial denaturation for 10 min at 94℃ for 1 cycle, then for 30 cycles at 94℃ for 1min, 60℃ for 1 min and 72℃ for 90 sec, final extension step for 5 min at 72℃ [11] by (Aeris Machine Peltier technology Thermo Assist). Then the amplified PCR products were analyzed using 1.5% Agarose gel with 30 μl ethidium bromide in electrophoresis along with a 100bp DNA ladder, by UV transilluminator system (Biometer an analytical Jena company) the gel visualized for presences of genes. Control; E.coli (ATCC 25922) was used as the negative control, PCR confirmed E.coli strain harboring ESBL genes were taken as positive control and nuclease-free waters as a negative control.
Data analysis
The Statistical package for social sciences (SPSS-version 21) performed and the significance of the p-value was tested by the chi-square test. In this cross-sectional study, SPSS was used to determine the frequency and percentage of antibiotic resistance-related genes.
Ethical consideration
Ethical Consideration: Well verbal informed the participant; as each patient participation was voluntary and anytime had the right to withdraw from the study, and the questionnaire and consent form of the participant's agreement was either sign or stamp (some patients uneducated) on the research tool on the complication of data collection, process, and publication. The research project approved by the research committee (SUMASRI-International Review Board); Faculty of Graduate Studies, University of Medical Sciences and Technology-Khartoum, Sudan.
Result
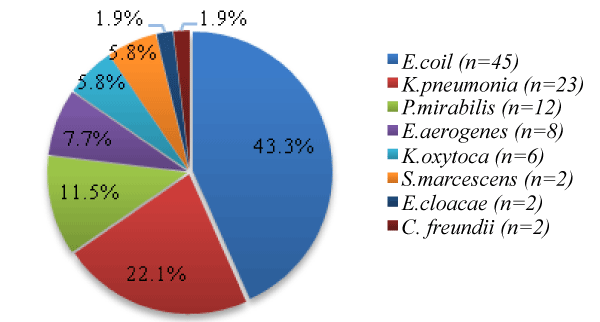
Hundred and four (n = 104) Enterobacteriaceae uropathogens isolated in (2007-2008), from midstream urine (MSU) samples. In 2019, only 56 (54%) of the recovered isolates were resistant to 3rd GCs, and of these 25 (44.6%), were ESBL-PE positive isolates. Figure 1 present the distribution of various Enterobacteriaceae uropathogens isolated species identified by API 20 E.
Antibiotic resistance pattern
The drug resistance patterns of 25 (44.6 %) uropathogenic of ESBL-PE positive isolates were observed as cefotaxime, ceftazidime, and co-trimoxazole showed the highest percentages (100%) of resistance among all other antibiotics by the disc diffusion method, followed by nitrofurantoin (92.0%), cefpodoxime and amikacin (72.0%) for each, ciprofloxacin (64.0%) and aztreonam (56.0%). The lowest rate of resistance was observed in gentamicin (36.0%) (Table 2).
Molecular screening of the ESBL-PE
The multiplex-PCR of the ESBL genes was performed to understand the most frequent ESBL genes in 2008 in our/region. Out of the 56 (54 %) 3rd GCs resistant Enterobacteriaceae isolates, 25 (44.6%) isolates harboured ESBL-PE genes. The predominating isolate was E. coli 15 (60.0%) followed by K. pneumoniae 9 (36.0%) and E.aerogenes 1(4%). The molecular characterization of the ESBL-PE is listed in Table 3. Molecular characterization of 25 ESBL-PE revealed that 10 (40 %) of them harboured blaTEM genes. Whereas blaCTX-M and blaSHV genes were detected in 4 (16.0%) and 2 (8.0%) of the isolates, respectively. The coexisted ESBL-PE was detected in 9(36 %) of the 3rd GC resistant uropathogens isolates, with (5/25; 20%) blaSHV/blaTEM, followed by (4/25; 16%) blaCTX-M/blaTEM. E. coli and K. pneumoniae uropathogens harboured all three resistance genes (blaTEM, blaCTX-M, and blaSHV), and E.aerogenes harboured only one gene (blaTEM) Figure 2. The Multiplex PCR for the three target ESBL genes is a significant method with a p-value of < 0.01 and chi-square statistic is 60.899.
Discussion
Regarding the spread of ESBL-PE in hospitals and their increased antibiotic resistance is alarming, though this situation was essentially unknown and still there is a snowballing of the antimicrobial resistance problem until now it is a critical problem in many countries. Consequently, the molecular characterization of beta-lactamases would be important for the reliable antimicrobial resistance gene identification besides an epidemical survey mainly among uropathogenic isolates as the UTI is a more common infection worldwide. The present retrospective study is on the distribution of uropathogenic ESBL-EP genes and their antimicrobial resistance pattern in the Sudanese population who were admitted to the urology department at ACTH.
The ESBL-PE antimicrobial resistance pattern in the study shows the resistance is high to commonly used antimicrobial agents used in UTI treatment policy, this finding is almost compared with similar studies around the world [12-17], and revealed that the resistance among ESBL-PE is higher than that of resistance in non-producers uropathogenic. Conversely, low incidences reported in other studies [18-20]. The frequency of resistance patterns among ESBLs-EP isolates can vary by country. It is observed that uropathogenic ESBL-EP isolates showed 100% resistance towards cefotaxime, ceftazidime, and co-trimoxazole, which is a much higher percentage when compared with others finding from [15,20]. Only 8.0% of the isolates were found susceptible to nitrofurantoin and therefore poorly effective. Nevertheless, aztreonam, ciprofloxacin, cefpodoxime, and amikacin can still be used as first-line therapy as considerable they have low resistance against those isolates tested.
The ESBLs producing gene distributions in the current study; blaTEM enzyme was predominant and the most common ESBL type at that time (40%) followed by blaCTX-M (20%) and blaSHV (8%) similar to those who reported [11,21,22]. The blaTEM variants were the most common ESBLs during the past decade, even though the identified numbers agree with findings in which ESBL enzymes of blaCTX-M and blaSHV type have been represented with a global- like spread pattern at research time [22-24]. Currently, the blaCTX-M genotypes have emerged as prevalent ESBL worldwide type, these increasing blaCTX-M frequency from other studies may be related to high mobilization of the encoding genes as it is transferred via plasmid, increased tenfold in comparison to other classes of beta-lactamase [25]. Concerning the new studies that were done in our region the most prevalent ESBL-gene is CTX-M type confirm that it is replacing SHV and TEM beta-lactamase enzymes theory [7,26,27]. Furthermore, there was an occurrence of ESBL co-producing genes reported in 9 strains of Enterobacteriaceae isolates were including mainly the blaTEM gene with either blaCTX-M or blaSHV, this result is in agreement with findings of [12,14,16,21,27,28] and is considered a frequent disseminated β-lactamases type. In our community, we need to address epidemiological studies and map the spread of antibiotic-resistant organisms throughout hospitals and in community, for a better update of infection prevention and control events.
Conclusion
The main finding of our study the blaTEM was the most prevalent β-lactamase followed by blaCTX-M and blaSHV genes produced by uropathogenic Enterobacteriaceae isolates from patients admitted to ATCH in Khartoum state, Sudan (2007-2008). The antimicrobial resistance pattern of ESBL-EP was higher towards cefotaxime, ceftazidime, co-trimoxazole, and nitrofurantoin, besides gentamicin, aztreonam and ciprofloxacin exhibited good in-vitro activity against uropathogenic ESBL-EP. The study highlights the importance of sequencing the entire blaTEM, blaSHV, and blaCTX-M gene types to track the clonal dissemination in our hospital and community for the evolution of the spread of beta-lactamase mediated resistance in the future studies.
Declarations
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
A deep appreciation extended thanks go to Dr. Khalid Enan (Central Laboratory Ministry of High Education and Scientific Research) for primers, PCR, and gel electrophoresis availability for work running 2019.
Funding
The research was fully funded by the 1st author in the frame of a master's degree submitted to the Faculty of Medical Laboratory Sciences at the University of Medical Sciences and Technology. No funding source.
Availability of data and materials
All relevant data is reported in the manuscript and attached document upon request.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
All authors read and approved the submitted version of this manuscript.
References
- Flores-Mireles AL, Walker JN, Caparon M, et al. (2015) Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13: 269-284.
- Foxman B (2013) Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28: 1-13.
- Khoshnood S, Heidary M, Mirnejad R, et al. (2017) Drug-resistant gram-negative uropathogens: A review. Biomed Pharmacother 94: 982-994.
- Shaikh S, Fatima J, Shakil S, et al. (2015) Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi journal of biological sciences 22: 90-101.
- Ahmed OB, Omar AO, Asghar AH, et al. (2013) Prevalence of TEM, SHV and CTX-M genes in Escherichia coli and Klebsiella spp Urinary Isolates from Sudan with confirmed ESBL phenotype. Life Sci J 10: 191-195.
- Bajpai T, Pandey M, Varma M, et al. (2017) Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J Med 7: 12.
- Abayneh M, Tesfaw G, Abdissa A (2018) Isolation of extended-spectrum ß-lactamase-(ESBL-) producing Escherichia coli and Klebsiella pneumoniae from patients with community-onset urinary tract infections in Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol 13: 2018.
- Kwon JH, Fausone MK, Du H, et al. (2012) Impact of laboratory-reported urine culture colony counts on the diagnosis and treatment of urinary tract infection for hospitalized patients. Am J Clin Pathol 137: 778-784.
- Hsueh P-R, Ko W-C, Wu J-J, et al. (2010) Consensus statement on the adherence to Clinical and Laboratory Standards Institute (CLSI) Antimicrobial Susceptibility Testing Guidelines (CLSI-2010 and CLSI-2010-update) for Enterobacteriaceae in clinical microbiology laboratories in Taiwan. J Microbiol Immunol Infect 43: 452-455.
- Pramanick D, Forstova J, Pivec L (1976) 4 M guanidine hydrochloride applied to the isolation of DNA from different sources. FEBS let 62: 81-84.
- Satir S, Elkhalifa A, Ali M, et al. (2016) Detection of Carbepenem resistance genes among selected Gram Negative bacteria isolated from patients in-Khartoum State, Sudan. Clin Microbiol 5: 266.
- Hassan H, Abdalhamid B (2014) Molecular characterization of extended-spectrum beta-lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Dev Ctries 8: 282-288.
- Higashino M, Murata M, Morinaga Y, et al. (2017) Fluoroquinolone resistance in extended-spectrum ß-lactamase-producing Klebsiella pneumoniae in a Japanese tertiary hospital: Silent shifting to CTX-M-15-producing K. pneumoniae. J Med Microbiol 66: 1476-1482.
- Ibrahim ME, Bilal NE, Magzoub MA, et al. (2013) Prevalence of extended-spectrum ß-lactamases-producing Escherichia coli from Hospitals in Khartoum State, Sudan. Oman Med J 28: 116.
- Mekki AH, Hassan AN, Elsayed DEM (2010) Extended spectrum beta lactamases among multi drug resistant Escherichia coli and Klebsiella species causing urinary tract infections in Khartoum. J Bacteriology Res 2: 18-21.
- Miftode E, Dorneanu O, Leca D, et al. (2010) Extended spectrum beta-lactamases in Escherichia coli and Klebsiella spp. from Eastern Romania. International Journal of Infectious Diseases 14: e39-e40.
- Sasirekha B, Shivakumar S (2012) Occurrence of plasmid-mediated AmpC ß-lactamases among Escherichia coli and Klebsiella pneumoniae clinical isolates in a tertiary care hospital in Bangalore. Indian J Microbiol 52: 174-179.
- Abujnah AA, Zorgani A, Sabri MA, et al. (2015) Multidrug resistance and extended-spectrum ß-lactamases genes among0 Escherichia coli from patients with urinary tract infections in Northwestern Libya. Libyan J Med 10: 26412.
- Aslan AT, Akova M (2019) Infections Caused by Extended-Spectrum ß-lactamase-Producing Enterobacteriaceae: Clinical and Molecular Epidemiology and Treatment Strategies. Infectious Diseases and Clinical Microbiology 1: 149-157.
- Mobaleghi J, Salimizand H, Beiranvand S, et al. (2012) Extended spectrum B-lactamases in urinary isolates of escherichia coli in five Iranian hospitals. Asian J Pharmaceutical and Clinical Res 5: 35-36.
- Seyedjavadi SS, Goudarzi M, Sabzehali F (2016) Relation between blaTEM, blaSHV and blaCTX-M genes and acute urinary tract infections. Journal of Acute Disease 5: 71-76.
- Shahid M, Singh A, Sobia F, et al. (2011) blaCTX-M, blaTEM, and blaSHV in Enterobacteriaceae from North-Indian tertiary hospital: high occurrence of combination genes. Asian Pacific Journal of Tropical Medicine 4: 101-105.
- Baraniak A, Grabowska A, Izdebski R, et al. (2011) Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob Agents Chemother 55: 5493-5499.
- Oteo J, Pérez-Vázquez M, Campos J (2010) Extended-spectrum ß-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis 23: 320-326.
- Zhao W-H, Hu Z-Q (2013) Epidemiology and genetics of CTX-M extended-spectrum ß-lactamases in Gram-negative bacteria. Crit Rev Microbiol 39: 79-101.
- Altayb HN, Siddig MA, El Amin NM, et al. (2018) Molecular Characterization of CTX-M ESBLs among Pathogenic Enterobacteriaceae isolated from different regions in Sudan. Global Advanced Research Journal of Microbiology (GARJM) 7: 40-47.
- Malik I, Elhag K (2019) Characterisation of Extended-Spectrum ß-Lactamases Among Multidrug Resistant Enterobacteriaceae From Sudan. J Pure Appl Microbiol 13: 61-68.
- Sharma M, Pathak S, Srivastava P (2013) Prevalence and antibiogram of Extended Spectrum ß-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diagn Res 7: 2173.
Corresponding Author
Magdi A Bayoumi, Faculty of Medicine, Department of Microbiology, University of Medical Sciences and Technology (UMST), Sudan.
Copyright
© 2021 Hamid OM. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.