DNA-Lipids Interactions in Nuclear Pore Assembly and Hormone Actions
Abstract
In a modern science there are ideas which were born earlier, than there was a necessary tool for their proof and development. Finally such ideas the oblivion waits, but it does not mean, that they are incorrect, simply scientific community has not ripened to their necessity, that result in scientific progress braking. The author shows on an example of one such idea: DNA-lipids interactions and DNA-membrane complexes. Having analyzed role of DNA-lipids complexes in vitro and in vivo we have proposed model of a nuclear pore which based on our understanding method such complexes assembly including the unwinding three-stranded DNA/RNA hybrid during pre-pore formation with ssDNA and double stranded DNA/RNA hybrid production. Interaction of nucleoporins with these polynucleotides in pore complex annulus leads to native structure of nuclear pore. The author propose new model of the first stages of a nuclear pore formation with participation three-stranded DNA/RNA hybrids and membrane vesicles without any proteins.
Keywords
DNA-lipids interaction, Nuclear pore, Vesicles, Hormones
Abbreviations
DMC: DNA-Membrane Complex; ssDNA: Single-stranded DNA; LMWRNA: Low Molecular Weight RNA
Introduction
There are a lot of ways to regulate genome activity at the level of transcription and most of them are related to DNA-protein interactions, where either transcription enzymes (RNA polymerase) or peptidic or protein hormones or huge other protein regulators of gene activity. The problem of structure and the role of DNA-lipid interactions, the existence of which was proven by studies conducted over the past 40 years are more complex as seemed to many. The biophysical and biochemical methods to study of DNA interaction with membranes, mainly with artificial ones (liposomes, Langmuir-Blodgett layers, etc.) used.
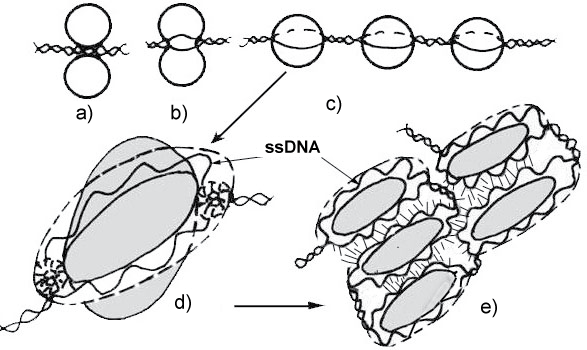
It is clear that the regulatory effect of some lipids on transcription processes is associated with the formation of DNA complexes with a membrane proteins or lipid bilayer. It is possible that some hormones that are hydrophobic in nature also affect DMC, changing the transcriptional properties of ssDNA (Figure 1).
Analyzing the properties of DNA-lipid complexes in vitro, the author proposed a model for the formation of such complexes in a cell and their possible participation in the formation of nuclear pores. It has been discovered recently that chromatin areas have an increased transcriptional activity near nuclear pores [1]. We believe that our model will well explain these regulatory functions of nuclear pores at the level of transcription. Our model also explains the structure of "transcription factories", which, in our opinion, are precursors of nuclear pores and also are a regulator of transcription in the nucleoplasm (Figure 1). Undoubtedly hormones should interact with both nuclear pores and with "transcription factories", which is currently shown by many authors, but the structure of such complexes is not clear [2]. Before explaining the mechanism of genome regulation through DMC, we should briefly state the data available today on the structure and functions of Ternary Complex (TC): DNA-zwitterionic liposomes-Mg++, nuclear pores and transcription factories. Then we try to introduce into their structure hormones and get a complete picture of the regulation by hormones cells activity.
DNA-Lipids Interactions In Vitro
Twenty years ago my attempts to have a talk about DNA-lipid interactions were usually accompanied with the question: "Whether such interaction exist really?"
As it is known in 1972 an Italian scientist Manzoli and his colleagues investigated DNA-lipid interactions in vitro and showed that this interaction was present. Depending on the type of lipid, DNA-lipid interaction is able to stabilize or destabilize DNA double helix [3,4]. A big progress in study of lipid-nucleic acid interactions has been made by Budker who described the formation of TC in 1978 [5-10]. Some features of DNA-lipid interactions are highlighted in our works [11-20].
The brief summary of the results of the study of ternary complexes is listed below:
DNA forms complexes with three main lipids: Phosphatidylcholine (PC); Phosphatidylethanolamine (PE) and Sphingomyelin (SM), the addition of other lipids increases or decreases these interactions force [12], Ability of divalent cations to form complexes with DNA and PC correlates with a degree of binding of these cations with PC [13], A cooperative character of binding of Mn2+ in the complex DNA-PC was shown [14], DNA is partially unwound in TC [5,15], DNA molecules in TC become restrictedly accessible to action of DNAse I, but are more accessible to digestion of S1-endonuclease [5], In addition to the double-stranded DNA, a three-stranded polynucleotide polyA*2polyU forms TC as revealed by microcalorimetry [16].
Double-stranded polynucleotides are able to absorb on the surface of-zwitterionic liposomes in the presence of divalent cations and also to induce liposome fusion (hemi-fusion). DNA-induced fusion or aggregation of liposomes depends on the liposomes size [17-20].
DNA-Membrane Complexes In Vivo
Observations of TC using an optical and an electron microscope revealed a huge diversity of structures formed by liposomes in TC. They are giant flat surfaces, sites of DNA unwinding on the surface of a lipid bilayer, bilayer fusion and hemi-fusion, etc. [21,22]. This promotes further search in the cell for similar structures which could be formed as a result of DNA-lipid interactions. Since chromatin DNA is positioned in the nucleus surrounded by nuclear envelope composed of lipids, 50%, then the desired structures (nuclear pores? pore complex lamina?) should be found at the periphery of chromatin which contacts with internal nuclear membrane. Therefore, we should consider the issue on DMC which was touched upon at the same time as DNA-lipid interactions in vitro study [22- 26].
In 1970-1980, an attempt was made to isolate a fraction of DMC and to analyze its biochemical composition. For this purpose, the cells have been gently lysed in presence of Mg2+ sarcosylate and so-called "M-band" fraction has been obtained. This fraction contained DNA, enriched by moderate repeats, newly synthesized DNA, RNA and proteins [24-26]. There was no information on lipid composition of "M-band" fraction. However, there is evidence on changes of lipid composition of nuclear envelope in cells, with increased level of transcription and the replication. This fact may suggest on important role of lipids in template synthesis of DNA and RNA [26].
The involvement of DMC in an initiation of replication and transcription and in a regulation of gene expression has been suggested [19,27,28].
Data on DNA-lipid interactions in vitro are beyond any doubt and they widen our knowledge in biophysics. DMC findings are thought to be an artifact since no appropriate techniques have been available to estimate their significance. In molecular biology the methods of PCR and DNA sequencing have not yet been developed at that period of time.
DNA-Membrane Complexes and Nuclear Pores
Now we may talk about interaction between DMC and nuclear pores. In 1968 Comings suggested that chromatin DNA is attached to the nuclear pores [21,22]. Modern interpretation of DNA attachment to nuclear pores through DNA-protein interactions appears to be unconvincing [29]. In the context of modern knowledge about the involvement of nuclear pores in transcription [30-32] this suggestion seems to be true since dense heterochromatin close to nuclear pores is diffuse, hence it is transcriptional active euchromatin. It is viewed in any electron microscopic images of the nucleus. Because of the absence of the biochemical techniques developed to date it was impossible for Comings to analyze a detailed DNA attachment to nuclear pores.
Biophysical study of TC showed that DNA in the course of TC formation is partially unwinding. Based on it the model of TC formation in vitro has been proposed and an attempt to explain the mechanism of nuclear pore formation with the involvement of DNA and membrane vesicles has been made [15,17,19,20]. In the course of study of TC the early model underwent changes and now it can be used to describe the formation of nuclear envelope from membrane vesicles and from endoplasmic reticulum too [33].
Simultaneously with our model of the formation of nuclear envelope and pore complexes with the involvement of lipids and chromatin DNA, the model of nucleoporin-based nuclear pores has been developed [34-36]. Although almost all nucleoporins are known, an attempt to build nuclear pore complex in vitro using only nucleoporins failed.
The use of our model provided us with the possibility to produce pore-like structures in lipid bilayer of liposomes which were added to the extract of Xenopus laevis eggs [37]. The annulate lamellae-like structures were also obtained. These structures are present in Xenopus eggs extract and very similar to nuclear pore complexes.
Are there irreconcilable contradictions between two approaches to study the formation of nuclear pores? Now we try to give an answer to the question concerning DMC. If DNA is bound to the pore complex, then why DNA is invisible in the area of nuclear pores in modern Electron Microscopy (EM) images of cellular nuclei? First, many scientists observed links between chromatin and nuclear pores [38,39]. Naked DNA is hardly visible even on the surface of liposome with the use of cryo-TEM (cryo-Transmission Electron Microscopy) high resolution method, but it is actually impossible to view it on the surface of nuclear membrane which contains proteins and lipids. At the same time cryo-TEM images are available where it is possible to view threads which extend from nuclear pore annuli inside nucleus.
It is difficult to select a dye for DNA for EM study since all dyes change DMC structure. Only staining with lanthanum and terbium which preferentially bind single-stranded DNA may make a situation clear. Staining with terbium of TC supports our model of TC where DNA double helix is unwinding in TC [18]. Nuclear pores are also stained with lanthanum [40,41].
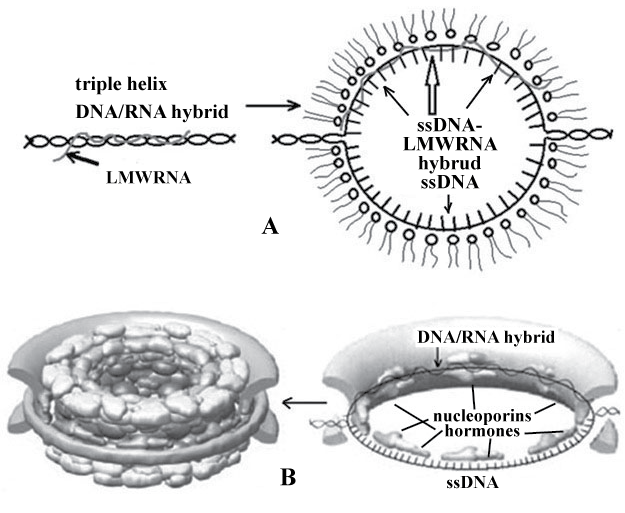
From our view the combination of our model of pore complexes and alternative models provides us with the possibility to produce nuclear pores in vitro system: DNA-liposomes-Mg2+ with the addition of nucleoporins or extract of Xenopus laevis eggs [38]. The modern model of nuclear pore taking into account the role of DNA and lipids is represented in Figure 2.
As it is seen ssDNA and DNA/RNA hybrid located in pore annulus were attached to Figure 2B. The model obtained seems not complex and has a series of advantages over the previous model. First, the presence of three-stranded DNA/RNA hybrids in chromatin induces fusion of membrane vesicles from which pre-pores are formed (Figure 1C and Figure 1D). Unwinding of DNA-RNA hybrid and the formation of single stranded DNA and LMWRRNA-DNA hybrid in pore annulus make annulus the site of nucleoporin attachment (FG-nucleoporins). Then other nucleoporins join them forming intranuclear and cytoplasmic surface of the pore (Figure 2B).
Thus, chromatin is attached to the nuclear envelope due to DNA-lipid interactions, which provides most strong binding of chromatin and the nuclear membrane, and nucleoporins.
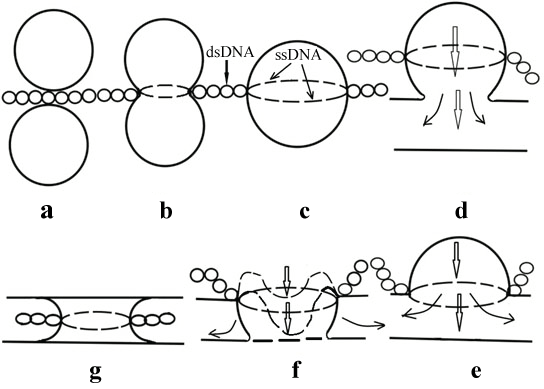
As follows from the above the issue on the existence of DMC is rather complicated. This issue can be referred to biophysics' one. The existence of DNA-lipid interactions in vitro is a proof of it [1-9]. We proposed several models of the formation of nuclear pores taking into consideration only DNA-lipid interactions [10,13,15,17,18]. The latest model of the formation of nuclear pores [41] even without the involvement of nucleoporins is rather complex and has a number of stages (Figure 3).
If these stages are excluded from consideration, then it is the model the majority of scientists study trying to build a pore complex using only nucleoporins. Probably it will be easier to obtain the desirable model using also all stages nuclear pore assembly shown on Figure 3. There are rather many facts which could not be ignored to support our model.
Conclusion
In conclusion, the problem of nuclear envelope formation requires not merely development of the existing approaches and inclusion of additional factors but complete change of the paradigm in this question. DNA-lipid interactions are currently the most obvious and deeply investigated area of biophysics [17] being in good agreement with the "nucleoporins approach" [41] to solution of the problem of nuclear pore formation.
The problem of nuclear pore formation involves the factors that cannot be overcome within the limits of the existing paradigm, e.g., the fusion of two very distant membranes with formation of a hole between them. There are numerous electron-microscopic observations of nuclear pore stages formation [42], that confirm our model and, as a consequence, the role of nuclear pores in transcription [43], which researchers try to solve in the framework of the nucleoporin model of the pores, whereas it can be solved only with our nuclear pore model.
Our model assumes the loop's organization of interphase chromatin and nucleoid. Such interphase chromatin organization gradually turn into the loop organization of mitotic chromosomes, and all this follows from acknowledgement of the real existence of DMC, which were thrown out of the arsenal of biology 30 years ago. Many problems of the nuclear matrix study can be explained on the basis of our DMC model and detailed analysis of the procedure of nuclear matrix isolation. The fact that nuclear pore formation involves hydrodynamic forces that appear during the DNA-induced fusion of cell membrane vesicles cannot be explained in principle within the limits of the nucleoporin's-based pore model. The time has come to combine our model with the dominant "nucleoporin model" of nuclear pores assembly for completion it.
Outlook
So, we got the latest idea about the structure of nuclear pores and should find a place for hormones in the pore structure. Since many of them are hydrophobic, it is logical to assume that hormones can interact with single-stranded DNA in a pore complex located in the lipid environment of the nuclear envelope. It can be assumed that the addition of certain hormones will positively affect on the transcription. It is logical to expect a similar mechanism of interaction of hormones with transcription factories; however, in this case the localization of the hormone will be intra-nuclear type [43]. There are hormones that inhibit transcription in nuclear pores, then they can be a good help in cancer treatment.
Accepting a new paradigm will result in increase of research activity in the field of TC, because the efforts of 4-5 laboratories engaged now in this problem worldwide are clearly insufficient. The author has succeeded, using an original technique, to isolate "membrane DNA", and there is a necessity of its further analysis including sequencing [44]. It should be acknowledged that the isolation and analysis of DMC and "membrane DNA" is a difficult problem, and we understand the previous unsuccessful attempts of its isolation [45]. The analysis of "membrane" ssDNA and LMW RNA sequence being a component of DMC will make it possible to analyze the most transcriptional active genes in the cell by analyzing the LMW RNA as part of pore complexes. The interaction between numerous nuclear receptors and DMC components will provide for a better understanding of their influence on genome expression.
Investigation of the role of TC in DMC and nuclear pore formation must also involve additional forces. The computer simulation of TC and first stages of nuclear pore assembly is the task of the nearest future. We have already spoken about the potential role of DMC in the processes of replication and transcription, hormone action, cancer progression and ageing of cells, as well as evolution of eukaryotes. The existing concept of the structure and functions of cell nucleus will be radically revised, what will result in the progress in treatment for many diseases that have been incurable up to now.
References
- Taddei A (2007) Active genes at the nuclear pore complex. Curr Opin Cell Biol 19: 305-310.
- Greenstein B, Wood D (2011) The Endocrine System at a Glance. (3rd edn), Wiley-Blackwell.
- Manzoli FA, Muchmore JH, Bonora B, et al. (1972) Interaction between sphingomyelin and DNA. Biochim Biophys Acta 277: 251-255.
- Manzoli FA, Muchmore JH, Capitani S, et al. (1974) Lipid-DNA interactions: II. Phospholipids, cholesterol, glycerophosphorylcholine, spingosine and fatty acids. Biochim Biophys Acta 340: 1-15.
- Budker VG, Godovikov AA, Naumova LP, et al. (1980) Interaction of polynucleotides with natural and model membranes. Nucleic Acids Res 8: 2499-2515.
- Gruzdev AD, Khramtsov VV, Weiner LM, et al. (1982) Fluorescence polarization study of the interaction of biopolymers with liposomes. FEBS Lett 137: 227-230.
- Mal'tseva TV, Bichenkov EE, Korobe A¬nicheva IK, et al. (1983) Interaction of poly A with phospholipid membranes using IR-spectroscopy. Biofizika 28: 766-770.
- Budker VG, Gorokhova OE, Sokolov AV (1984) DNA translocation across model phospholipid membranes. Dokl Akad Nauk SSSR 278: 479-482.
- Savchenko EV, Korobeinicheva IK, Budker VG (1985) Study of poly(C) and poly(I)-poly(C) complexes with model phospholipid membranes by infrared spectroscopy. Biofizika 30: 844-848.
- Budker VG, Degtiarev SKh, Sokolov AV (1986) Cleavage of DNA adsorbed on the surface of phospholipid membranes by restrictases type II. Biokhimiia 51: 1496-1498.
- Shabarshina LI, Sukhorukov BI, Kuvichkin VV (1979) Infrared spectroscopic study of DNA- lipid interactions. DNA compacting on disperse particles. Biofizika 24: 990-994.
- Kuvichkin VV (1983) Theoretical model of DNA-membrane contacts. Biofizika 28: 771-775.
- Kuvichkin VV, Sukhomudrenko AG (1987) Interaction of natural and synthetic polynucleotides with liposomes in the presence of divalent cations. Biofizika 32: 628-633.
- Kuvichkin VV, Volkova LA, Naryshkina EP, et al. (1989) 31PNMR and Mn2+ -ESR study of complexes of DNA-liposomes. Biofizika 34: 405-409.
- Kuvichkin VV (1990) Ultrastructural study of DNA-liposomes-Mg2+ complexes. Biofizika 35: 256-262.
- Kuvichkin VV, Kuznetsova SM, Emeljanenko VI, et al. (1999) Calorimetric study of the complexes: polyApolyU-phosphatidylcholine liposomes-Mg2+. Biofizika 44: 430-435.
- Kuvichkin VV, Danev RS, Shigematsu H, et al. (2009) DNA-Induced Aggregation and Fusion of Phosphatidylcholine Liposomes in the Presence of Multivalent Cations Observed by the Cryo-TEM Technique. J Membr Biol 227: 95-103.
- Kuvichkin VV (2009) Investigation of Ternary Complexes: DNA-Phosphatidylcholine Liposomes-Mg2+ by Freeze-Fracture Method and Their Role in the Formation of Some Cell Structures. J Membr Biol 231: 29-34.
- Kuvichkin VV (2002) DNA-lipid interactions in vitro and in vivo. Bioelectrochemistry 58: 3-12.
- Kuvichkin VV (2010) Lipid-nucleic acid interactions as base organization and expression of cellular genome. Int J Quant Chem 110: 120-126.
- Zhanga B, Chena Y, Hana Z, et al. (1998) The role of keratin filaments during nuclear envelope reassembly in Xenopus egg extracts. FEBS Lett 428: 52-56.
- Comings DE (1968) The rationale for an ordered arrangement of chromatin in the interphase nucleus. Am J Hum Genet 20: 440-460.
- Comings DE, Okada TA (1973) DNA replication and the nuclear membrane. Journal of Molecular Biology 75: 609-618.
- Harmon JM, Taber HW (1977) Some properties of a membrane-deoxyribonucleic acid complex isolated from Bacillus subtilis. J Bacteriol 129: 789-795.
- Hobart P, Duncan R, Infante AA (1977) Association of DNA synthesis with the nuclear membrane in sea urchin embryos. Nature 267: 542-544.
- Albi E, Viola Magni MP (2004) The role of intranuclear lipids. Biol Cell 96: 657-667.
- Kuvichkin VV (1982) The role of membrane in organization and gene expression of procaryotes and eucaryotes. In: The DNA-membrane complexes: data about their structure and functions, Moscow.
- Kuvichkin VV (1982) The role of membrane in organization and gene expression of procaryotes and eucariotes. In: Quantitative model of DNA-membrane contacts. Moscow, 72.
- Brown CR, Silver PA (2007) Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev 17: 100-106.
- Kalverda B, Pickersgill H, Shloma VV, et al. (2010) Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360-371.
- Van de Vosse DW, Wan Y, Wozniak RW, et al. (2011) Role of the nuclear envelope in genome organization and gene expression. Wiley Interdiscip Rev Syst Biol Med 3: 147-166.
- Mendjan S, Taipale M, Kind J, et al. (2006) Nuclear Pore Components Are Involved in the Transcriptional Regulation of Dosage Compensation in Drosophila. Mol Cell 21: 811-823.
- Anderson DJ, Hetzer MW (2007) Shaping the endoplasmic reticulum into the nuclear envelope. J Cell Science 121: 137-142.
- D'Angelo MA, Hetzer MW (2008) Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18: 456-466.
- Hoelz A, Debler EW, Blobel G (2011) The structure of the nuclear pore complex. Annu Rev Biochem 80: 613-643.
- Grossman E, Medalia O, Zwerger M (2012) Functional architecture of the nuclear pore complex. Annu Rev Biophys 41: 557-584.
- Kuvichkin VV, Uteshev VK (2003) The role of lipid-nucleic acid interactions in the reconstruction in vitro of the nuclear shell with pores. Biofizika 48: 236-239.
- Arlucea J, Andrade R, Alonso R, et al. (1998) The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J Struct Biol 124: 51-58.
- Ishii K, Arib G, Lin C, et al. (2002) Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551-562.
- Shaklai M, Tavassoli M (1982) Preferential localization of lanthanum to nuclear-pore complexes. Journal of Ultrastructure Res 81: 139-144.
- Park PC, De Boni U (1992) Nuclear membrane modifications in polytene nuclei of Drosophila melanogaster. Serial reconstruction and cytochemistry. Anatom Rec 234: 15-26.
- Kuvichkin VV (2011) The mechanism of a nuclear pore assembly: a molecular biophysics view. J Membr Biol 241: 109-116.
- Wu Y, Koenig RJ (2000) Gene regulation by thyroid hormone. Trends Endocrinol Metab 11: 207-211.
- Nagy V, Hsia KC, Debler EW, et al. (2009) Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. PNAS 106: 17693-17698.
- Kiseleva E, Rutherford S, Cotter LM, et al. (2001) Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J Cell Sci 114: 3607-3618.
Corresponding Author
Kuvichkin VV, Department of Reception Mechanisms, Institute of Cell Biophysics, Russian Academy of Sciences, 142290, Pushchino, Moscow Region, Russia, Tel: +79166405267.
Copyright
© 2017 Kuvichkin VV. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.