Chromosome Counting in Few Species of Musa by Traditional Technique
Abstract
The determination of ploidy level/chromosome numbers in different germplasms is important for better management of genetic diversity and is required before starting concerted breeding programs. The number of chromosomes sets in a cell is one of the defining characters of a species or cultivar. In the present study chromosome counting of Musa spp. were carried out by traditional method and revealed that Elachi trait had 11-12 chromosomes per cell, whereas, in the Poovan, 13-14 chromosomes per cell were seen and in Nendran, about 10-11 chromosomes per cell were counted.
Keywords
Banana, Traits, Chromosome
Introduction
Banana (Musa sp.) belongs to the family Musaceae and is among the tallest monocotyledons. Banana is one of the oldest fruit known to mankind and also important food for man. Commonly known as 'Apple of Paradise'. It is one of the earliest crops cultivated by man and still remains as the major staple fruit crop for millions of people in the tropical world. It constitutes an important component of the diet of rural and working-class people, as the fourth important global commodity in terms of the gross value of produce after rice, wheat and maize/milk products [1] and an important source of high calorific energy [Rich source of energy (137 K. Ca/100 g)] and a good laxative. It is not only the source of easily digestible carbohydrates but also provides essential vitamins B, C and several minerals such as potassium, calcium, and magnesium and has several medicinal properties. India is the largest producer of banana in the world, contributing 24% to the global production of banana, with a total production of 19.19 million tonnes from 0.565 million hectares. Brazil is the next largest producer of banana followed by the Philippine, Indonesia, China, and Ecuador. Banana is well adopted in India in the region's varying from humid tropical and semi-arid subtropics, and from the sea level to an elevation of 2,000 meters above mean sea level [2].
Ploidy is the number of complete sets of chromosomes in a cell. Somatic cells, tissues, and individuals can be described according to the number of sets of chromosomes present per cell (i.e. the Ploidy level). The number of chromosomes sets in a cell is one of the defining characters of a species or cultivar. Wild species and subspecies of banana are all diploid, whereas cultivars can be diploid, triploid or tetraploid. Ploidy has conventionally been determined by counting chromosomes in dividing root tip cells, a labor-intensive procedure that is made difficult in Musa by the fact that the chromosomes are small and numerous. The traditional method of chromosome counting in Musa spp. is being done by manually counting the number of chromosomes in the actively dividing metaphase cells under higher magnification of about 1000 X. The importance of morphology and number of chromosomes in Musa spp. is useful in the characteristic of a species. More detailed cytological studies are complicated by the small size of chromosomes. The aim of this work was to the counting chromosomes in Musa spp. (Elachi, Poovan and Nendran) by traditional techniques and it is useful for the characteristic of a species.
Materials and Methods
Sample collection
The essential part of this experiment i.e., roots were collected from the Musa spp. by removing underground roots and small roots having the small root tips of about 1 cm are excised and brought to the laboratory for further studies in the zip-lock polythene bags. For the present work three commonly available traits of Musa were selected they are, Elachi, Poovan, and Nendran Banana traits.
Storage of collected materials
The collected root tips were directly subjected to treated with 0.03% 8-hydroxyquinoline for about 4-9 hours and then the roots are transferred to the maceration fixation solution/storing solution for minimum 12 hours before the slide preparation of the collected materials. The roots may be stored in the same solution until further use.
Root sampling and treatment
The collected root tips (1 cm long) immerse directly in 1 ml of 0.03% 8-hydroxyquinoline solution at 20-22 °C for 4 to 9 hours. Transfer the root tips into the maceration fixation solution in sealed tubes (5-10 ml) for about 12 hours (overnight) to get the macerated tissues.
Slide preparation
From the maceration fixation solution pull out macerated tissues of the root tip and briefly blot dry on filter paper with the forceps. Take the apical root portion of these tissues approximately about 1.5 mm length and place the apical portion in the center of a microscope slide in a drop of the staining solution. Firmly squash the preparation on a slide with the help of a finger without horizontal moving. Remove the excess stain off the slide with filter paper. Flatten the preparation with radial vertical pressures from the center to the exterior of the slide with the eraser of the pencil. Pressure must be exercised to remove air bubbles from the preparation with enough force to spread chromosomes within the cells but not to burst cell walls. Remove excess of stain on the slide with filter paper. Finally, wait for 6 to 8 hours before starting the observations. The stain should take about two days to intensify and provide good contrast. The preparations should also be sealed with nail polish for later microscopic observation.
Observation of useful cells
In a clean slide make a mark on the top right of the slide to identify the observing materials on the slide. Observe the slides under the microscope. Locate the position of all good cells with well-identified chromosomes on the slide. Complete the scan of the slide and position the previously identified good cells by the noted coordinates. Pass the microscope to the 100X objective and observe the cells to count the chromosomes.
Chromosome counts on useful cells
Select the sites show only the cells with the chromosomes well separated and little overlap, in which chromosomes can be unequivocally defined and counted. Make a drawing and a photograph or an electronic image of the cell showing all the chromosomes then count chromosomes from five or six independent cells from the same plant material.
Results and Discussion
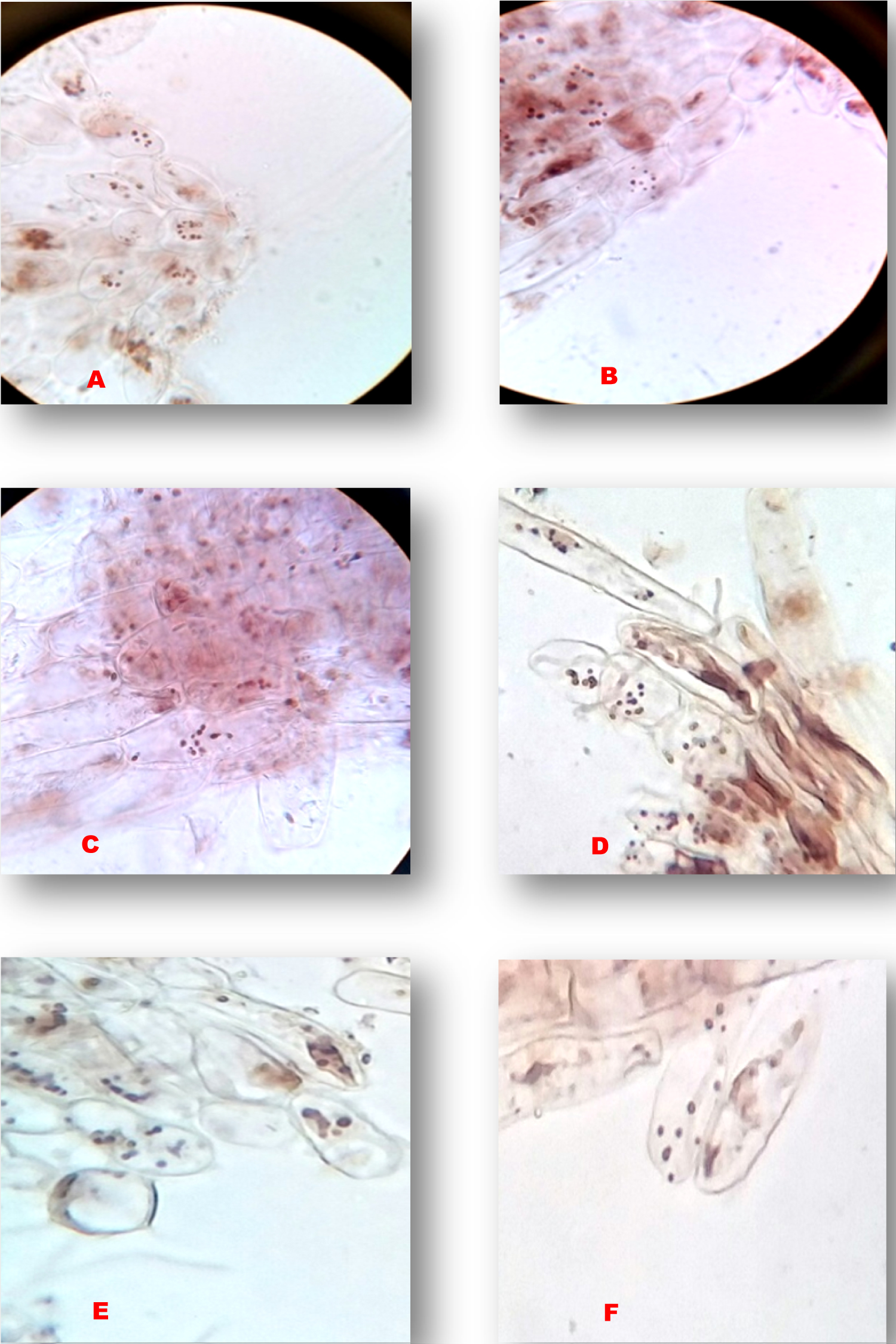
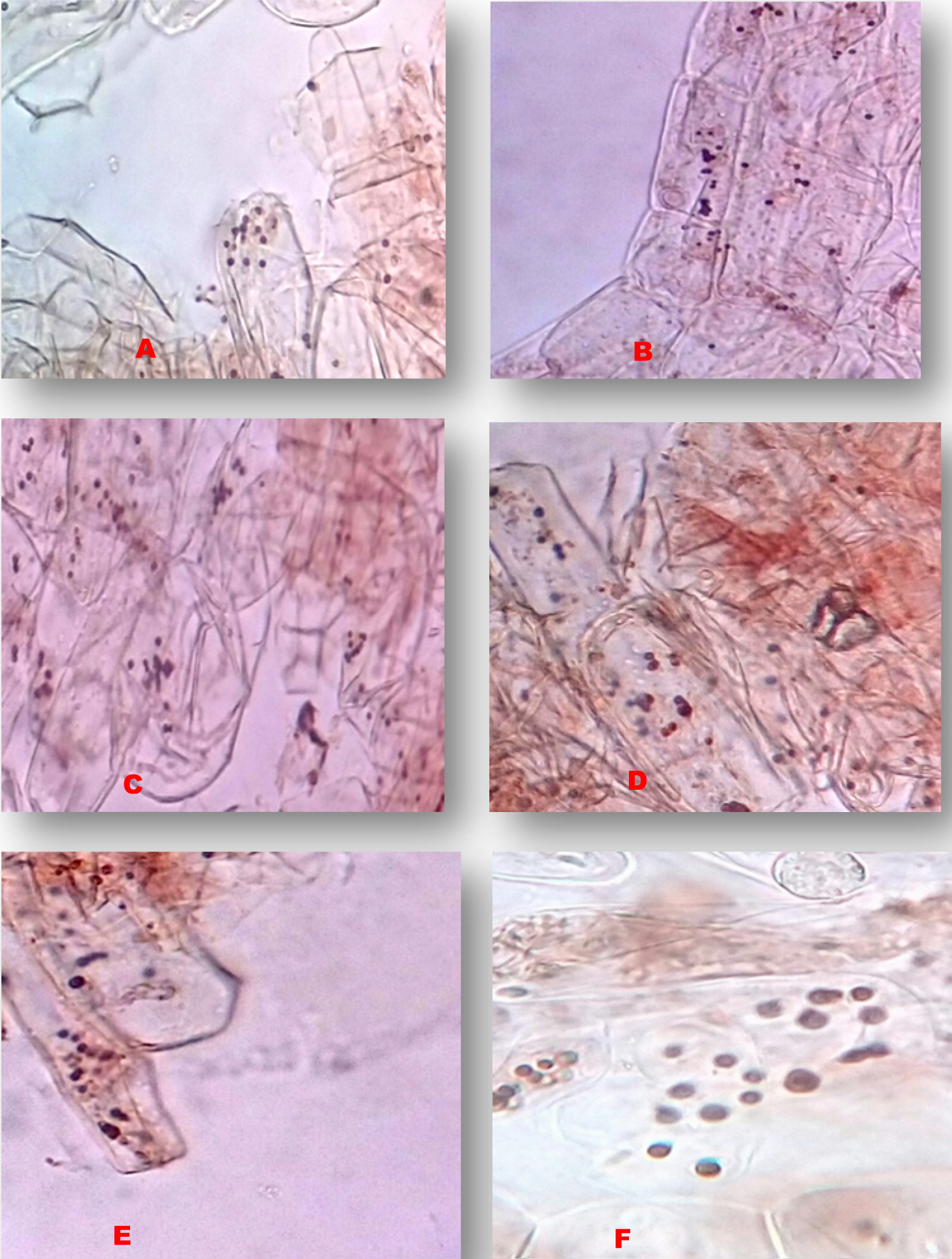
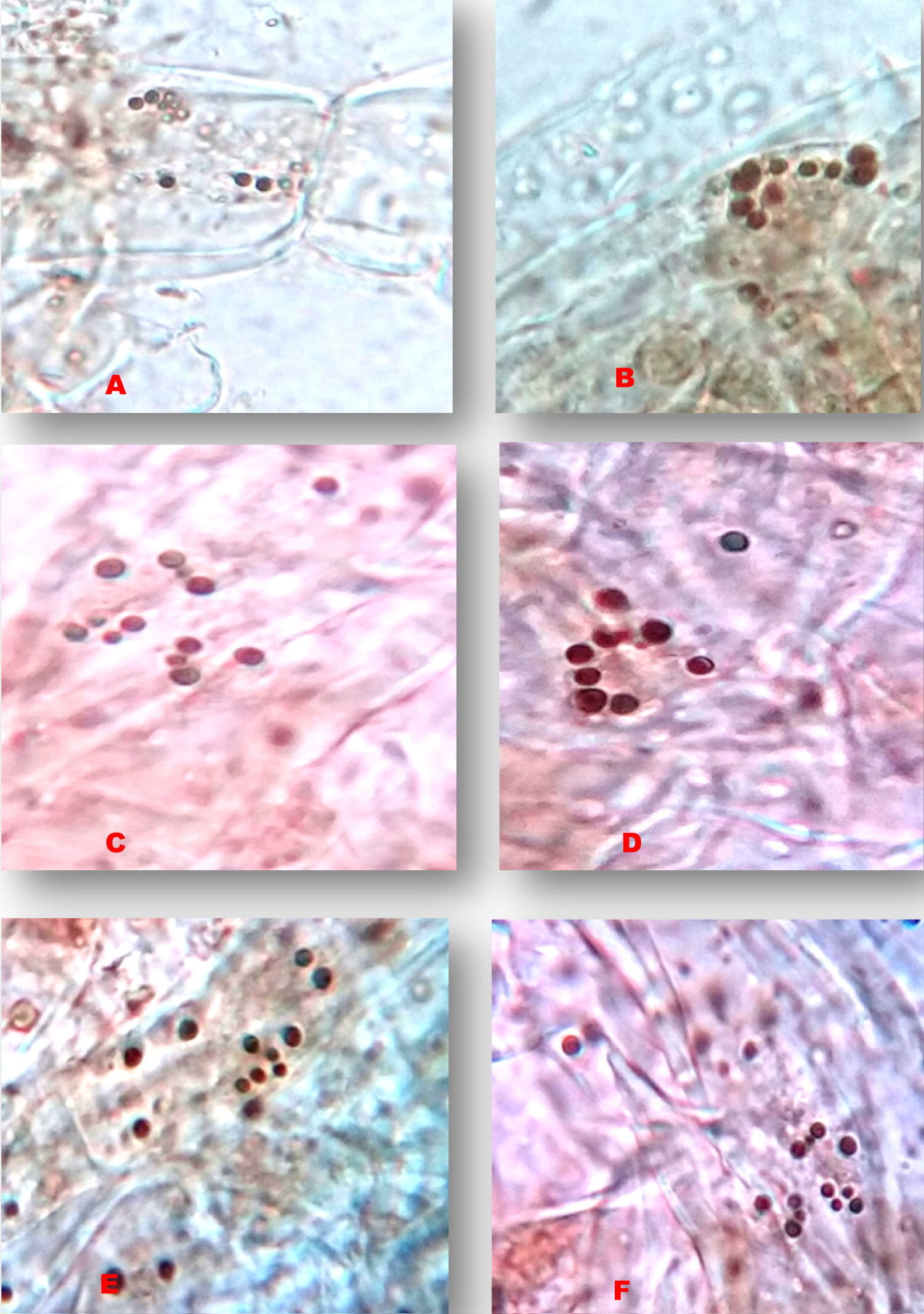
The chromosome counting of Elachi, Poovan and Nendran Banana traits, the following results were recorded in Table 1. In the traditional technique of the chromosome counting, 10 microscopic fields of well-spread cells were taken into consideration (Figure 1, Figure 2 and Figure 3). In the Elachi trait, 11-12 chromosomes per cell are being observed (Figure 1), whereas in the Poovan, 13-14 chromosomes per cell were seen (Figure 2). In Nendran, about 10-11 chromosomes per cell were counted (Figure 3). On an average, number of chromosomes recorded in Poovan (13.2), followed by Elachi with 11.1 chromosomes and Nendran with 10.9 chromosomes.
In the genus Musa, accurate determination of the ploidy by chromosome counting is laborious [3]. Agrawal [4] revealed cytological abnormalities of 17 South Indian banana varieties. Chromosome counts of 20 Bihar banana cultivars were reported by Roy and Sharma [5]. Valsala and Nair in 1990 reported a chromosome number of 98 cultivars [6]. Somatic chromosomes of 53 Musa landraces and hybrids were reported by Osuji, et al. [7] and 16 Musa species and landraces by Dolezel, et al. [8]. Variations were observed within groups may have resulted in the evolution of new cultivars. In the present investigation, the Bakry and Shepherd [9] protocol was followed for the counting of chromosome numbers. Chromosome counting methods are a basic tool for the determination of ploidy in Banana. Very less work had been carried out about counting of chromosomes in Banana by traditional techniques.
References
- Frison EA, Sharrock SL (1999) The economic, nutritional and social importance of bananas in the world. In: C Picq, E Foure´ and EA Frison, Bananas and Food Security, International Symposium, Douala, Cameroon, 21-35.
- Suman S, Rajak KK, Kumar H (2012) The diversity of genome and Ploidy in banana and their effect on tissue culture responses. Research in Environment and Life Sciences 5: 181-183.
- Nsabimana A, Staden J (2006) Ploidy investigation of bananas (Musa spp.) from the National Banana Germplasm Collection at Rubona-Rwanda by flow cytometry. South African Journal of Botany 72: 302-305.
- Agrawal PK (1983) Cytogenetical investigations in Musaceae I Meiotic Studies in South Indian bananas. Cytologia 48: 847-852.
- Roy RS, Sharma G (1951) Chromosome studies of Bihar bananas. Ind J Genetics Pl Breeding 11: 211-214.
- Valsala KPK, Nair PCS (1990) Cytotaxonomical studies on banana cultivars. South Indian Hortl 38: 177-182.
- Osuji JO, Okoli BE, Oritz R (1996) An improved procedure for mitotic studies of the Eumusa section of the genus Musa L (Musaceae). Infomusa 5: 12-14.
- Dolezel J, Dolezelova M, Roux N, et al. (1998) A novel method to prepare slides for higher solution chromosome studies in Musa spp. Infomusa 7: 3-4.
- Bakry F, Shepherd K (2008) Chromosome count on Banana root tip squashes. Banana Protocol Fruits 63: 179-181.
Corresponding Author
Veerabhadra Swamy AL, Assistant Professor, Post Graduate Department of Botany, J.S.S. College of Arts, Commerceand Science, B.N. Road, Mysore - 570 025, Karnataka, India
Copyright
© 2020 Veerabhadra SAL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.