Cloning and Identification of an Ubiquitin-Conjugating Enzyme E2 N Gene from Lethenteron Camtschaticum
Abstract
The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. Ubiquitin-conjugating enzyme E2N is a protein that is encoded by the UBE2N gene. Here, we report an Ubiquitin-conjugating enzyme UBE2N (LaUBE2N) gene in lamprey, which contained 456-bp Open Reading Frame (ORF) encoding 152 amino acids with a typical UBC domain. Real-time PCR assay showed that the highest expression of the protein in adult lamprey was in the leukocytes, the lowest expression was in the skin, kidney and liver. The high conservation in amino acid sequence of the LaUBE2N protein with the UBE2Ns from Homo sapiens, Ovis aries, Taeniopygia guttata and Danio rerio, implied that it had similar function with UBE2N proteins from other species.
Keywords
Ubiquitin-conjugating enzymes, UBE2N, Ubiquitin, Evolution, Lamprey
Introduction
Protein ubiquitination is mediated by the sequential action of an Ubiquitin-Activating Enzymes (E1s), an Ubiquitin-conjugating enzymes (E2s), and a range of Ubiquitin-Protein Ligases (E3s) [1-3]. E2s has fostered important insights into ubiquitin transfer mechanisms. Ubiquitin-conjugating enzyme E2 N (UBE2N) is a protein that in humans is encoded by the UBE2N gene, also known as Ubc13, UbcH13, Ubiquitin carrier protein N, or Ubiquitin-protein ligase N. UBE2V2-UBE2N heterodimer can interact with the E3 ligase STUB1/CHIP (via the U-box domain) and the complex has a specific 'Lys-63'-linked polyubiquitination activity [4,5]. The human RING Finger (RNF) E3 ubiquitin ligases, RNF8 and RNF168, with the E2 ubiquitin-conjugating complex Ubc13 (UBE2N)/UBE2V2 (Mms2), perform the majority of Lys-63 ubiquitylation in homologous recombination. The crystal structure of the RNF8/Ubc13/Mms2 ternary complex reveals the structural basis for the interaction between Ubc13 and the RNF8 RING in the DNA damage response, which is subsequently amplified by RNF168 [6]. The interactions of UBE2N with the E3 ligases, HLTF and SHPRH promote the 'Lys-63'-linked polyubiquitination of PCNA upon genotoxic stress and lead to DNA repair [7-9]. UBE2N also interacts with OTUB1 and interfere with UEV1A (also known as UBE2V1) binding to UBC13. OTUB1 binding also occludes the RING E3 binding site on UBC13 and leads to inhibit E2-conjugating activity [10-13].
By now, about ubiquitin proteasome system in lamprey was known little. In this study, we described the identification and cloning of a Ubiquitin-conjugating enzyme UBE2N gene from the leukocyte of Lethenteron camtschaticum, named LaUBE2N. The LaUBE2N could well be the mammalian counterpart, which might play important roles in ubiquitin proteasome pathway of lamprey lymphocyte.
Material and Methods
Animals
Adult Lethenteron camtschaticum (male and female, about 48-60 cm in length) were obtained from the Heilongjiang River region of Heilongjiang province, China. These lampreys were kept in Fiber Reinforced Plastic (FRP) tanks with freshwater at 4~16 ℃ before the experiments.
Separation of lamprey leukocytes
Peripheral blood was collected from the caudal subcutaneous sinus of the animal, and the leukocytes were enriched by Ficoll-Paque gradient centrifugation. In brief, the peripheral blood was first diluted with two volumes of D-Hanks, and the mixture was slowly applied onto the surface of the Ficoll-Paque medium (1.092 g/ml) in a centrifuge tube. After centrifugation at 160 × g for 10 min, the band formed at the interface between the Ficoll-Paque and the overlying solution, which consisted of the cells, was collected and diluted with another five volumes of D-Hanks followed by further centrifugation. The precipitate at the bottom of the tube was collected and preserved at -80 ℃ in cryoprotectant containing 10% DMSO and 90% FBS for real-time PCR analysis.
Cloning of Ubiquitin-conjugating enzyme LaUBE2N cDNA
Unless specified, the enzymes, cloning kits and vectors used in this study were all purchased from TaKaRa Bio Inc. (http://www.takara-bio.com/) the total RNAs of the leukocytes were extracted with RNA isolation kit, and then converted into complementary cDNAs with reverse transcriptase in TaKaRa cDNA Library Construction Kit. The cDNA were cloned into a pAP3neo Vector and sequenced. Based on the analysis of leukocyte ESTs library, an Ubiquitin-conjugating enzyme UBE2N homologs in Lethenteron camtschaticum was confirmed with the aid of Basic Local Alignment Search Tool X (BLASTx) in the National Center for Biotechnology Information (NCBI). A fragment of LaUBE2N cDNA was obtained by further sequencing using the cDNA Library plasmid.
Amino acid sequence analysis and phylogenetic analysis
The amino acid sequence of protein was analyzed at http://blast.ncbi.nlm.nih.gov/Blast.cgi and http://www.uniprot.org/uniprot. Multiple sequences of UBE2 were obtained from GenBank (https://www.ncbi.nlm.nih.gov/) and aligned using GeneDoc software. Protein structure prediction was analyzed by Protein Homology/analogY Recognition Engine V 2.0 on the web (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The MEGA 4 program was used to construct phylogenetic trees and the evolutionary history was inferred using the Neighbor-Joining method. A bootstrap analysis was performed using maximum parsimony as criteria for getting the optimal tree. The sequence of LaUBE2N gene identified in Lethenteron camtschaticum had been submitted to GenBank sequence database.
Real-time PCR
Total RNAs from different lamprey tissues (including whole blood cells, leukocytes, gill, liver, heart, kidney and intestine) were separately isolated using the Trizol reagent (Invitrogen, USA), and then subjected to reverse transcription using the Prime ScriptTM RT reagent Kit (TaKaRa, Japan) according to the manufacturer's instruction. Quantifications of UBE2N mRNA expression in different tissues were performed with a TaKaRa PCR Thermal Cycler Dice Real Time System. The GAPDH gene of Lethenteron camtschaticum was used as an internal control. The level of BTK-like mRNA was calculated relative to the amount of GAPDH mRNA. All primer sequences are listed in Table 1. The following PCR program for LaUBE2N was used: 30 s at 95 ℃, 40 cycles of 5 s at 95 ℃ 10 s at 52 ℃ and 15 s at 72 ℃. Subsequently, a melting curve analysis was performed, which consisted of 70 cycles of 10 s with a temperature increment of 0.5 ℃ per cycle starting at 56 ℃. The Cycle Threshold (Ct) values were obtained. Each sample was analyzed in triplicate. Data were analyzed with the SAS Proprietary Software Release 8.02.
Results
Identification of LaUBE2N gene in Lethenteron camtschaticum
A single EST sequence homologous to UBE2N was found among the extensive EST sequences from the cDNA library of Lethenteron camtschaticum blood cells previously constructed by our laboratory. By sequencing the plasmid in the cDNA library, a fragment of LaUBE2N cDNA with 641 nucleotides was obtained, which contained 456-bp Open Reading Frame (ORF) encoding a polypeptide of 152 amino acids (Figure 1A). The amino acids sequence from 6aa to 149aa was catalytic domain homologues of Ubiquitin-conjugating enzyme E2. The LaUBE2N cDNA sequences were submitted to GenBank database with the accession number KP203889.
The deduced amino acid sequence of LaUBE2N cDNA had high sequence identity with the UBE2Ns from other vertebrates (97% identity with Homo sapiens, 96% identity with Ovis aries and Taeniopygia guttata and 94% identity with Danio rerio) (Figure 2 and Supplementary Table 1). The high conservation in amino acid sequence of the LaUBE2N protein implied that it had similar function with UBE2N proteins from other species.
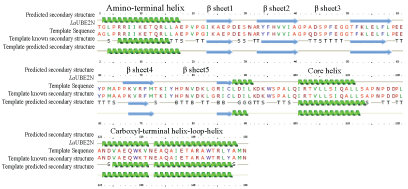
Protein structure prediction on the web using the Phyre server showed that LaUBE2N fold featured a five-strand antiparallel β sheet into which intercalates a helix to form a hydrophobic core. The hydrophobic core is flanked by an amino-terminal a helix and a carboxyl-terminal helix-loop-helix (Figure 3).
Phylogenetic analysis of LaUBE2N cDNA
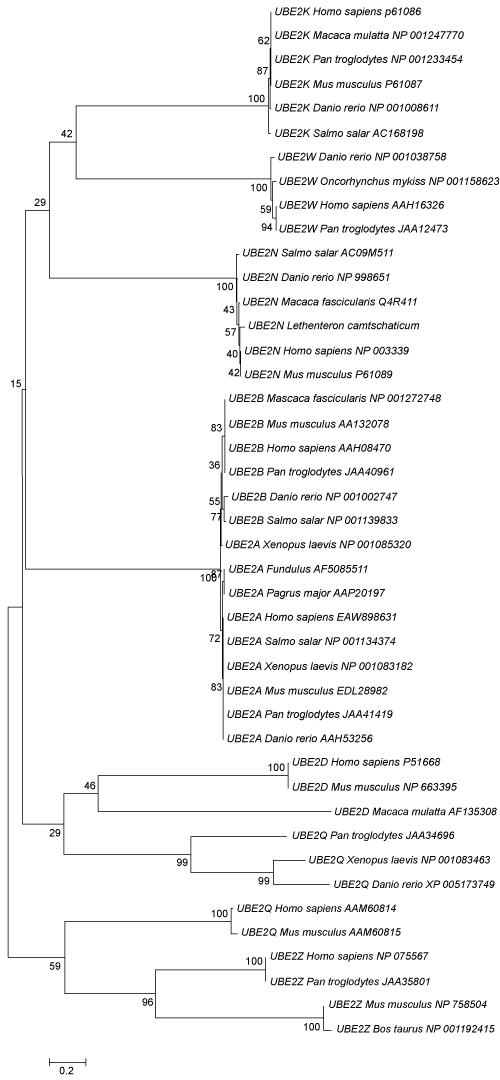
Neighbour-Joining phylogenetic trees showed that LaUBE2N sequence was clustered into UBE2N group (Figure 4).
Expression pattern of LaUBE2N in adult tissues
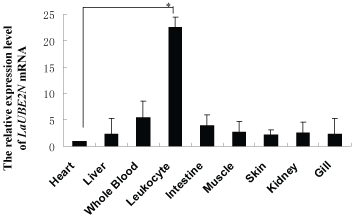
The expression of the LaUBE2N gene in various Lethenteron camtschaticum tissues was examined using Real-time PCR. The level of LaUBE2N mRNA was highest in the leukocyte, while the rest of the tissues had much lower levels (Figure 5).
Discussion
Lamprey and hagfish are surviving representatives of some of the most ancient cyclostomes, and are thought to have existed largely unchanged for more than 500 million years [14,15]. Features studies of lamprey have provided information on the origin and evolution of hallmark vertebrate characteristics such as the neural crest, ectodermal placodes, and jaw. In addition, these eel-like fishes use Leucine-Rich-Repeat (LRR)-based receptors, called Variable Lymphocyte Receptors (VLRs) for antigen recognition instead of the Immunoglobulin (Ig)-based receptors used by vertebrates, which provide insights into mechanisms of parallel evolution of adaptive immune response system [16-18]. So, lamprey has been poised to become an evolutionary developmental model of choice. Here we report for the first time, the identification of a novel UBE2N gene in Lethenteron camtschaticum. The fragment of LaUBE2N cDNA was obtained from leukocytes cDNA library of Lethenteron camtschaticum. The deduced amino acid sequence of LaUBE2N showed a typical catalytic domain homologue of Ubiquitin-conjugating enzyme E2. it is well known that there are over 30 E2 enzymes in humans, which are broadly grouped into four classes, all of which have a core catalytic domain (containing the active site cysteine), and some of which have short N- and C-terminal amino acid extensions: class I enzymes consist of just the Catalytic Core Domain (UBC), class II possess a UBC and a C-terminal extension, class III possess a UBC and an N-terminal extension, and class IV possess a UBC and both N- and C-terminal extensions [19]. These extensions appear to be important for some subfamily function, including E2 distribution and protein-protein interactions. In addition, there are proteins with an E2-like structure that are devoid of catalytic activity, but which appear to assist in poly-ubiquitin chain formation. LaUBE2N has UBC domain and both N- and C-terminal extensions and belongs to class IV.
Sequence conservation within the catalytic core domain of the E2 superfamily is reflected in a common E2 folding motif [20-22]. The hydrophobic core of UBE2N formed by a five-strand anti-parallel β sheet with a helix reflected shared requirements for binding El ternary complex. The helix-loop-helix is required for E3α binding and that the triad serves to anchor this segment to the hydrophobic core. Though, the El and E3α of lamprey were not reported by now.
To further analyze the relationship between Lethenteron camtschaticum UBE2N protein and UBE2Ns from other species, a phylogenetic tree was constructed, and the result of which suggested that the ancestor of UBE2N was present from agnatha vertebral animals to mammal. The similar structure between Lethenteron camtschaticum UBE2N protein and mammal UBE2Ns suggested that they may have similar functions. In addition, the amino acids sequence of LaUBE2N protein has higher homology with those of other species.
Lethenteron camtschaticum UBE2N gene was expressed in various tissues, yielding the highest expression in the leukocytes, the lowest expression in the kidney, heat, intestine, whole blood, muscle, skin and liver, which implied that LaUBE2N play a key role in leukocytes.
In conclusion, we identified a member of the Ubiquitin-conjugating enzyme 2 in lamprey, named LaUBE2N, which is of typical features of catalytic domain homologue of Ubiquitin-conjugating enzyme E2 proteins. It was expressed in various tissues including leukocytes, muscle, gill, heart, skin, intestine and liver. About the exact function of LaUBE2N need to be further studied.
Acknowledgments
This work was supported by grants from doctor initial foundation of Guangdong Medical College (No. XB1362) and Postdoctoral Science Foundation of China (No. 2013M530384).
References
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373-428.
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425-479.
- Roos-Mattjus, P Sistonen L (2004) The ubiquitin-proteasome pathway. Annu Rev Med 36: 285-295.
- Slotman JA, da Silva Almeida AC, Hassink GC, et al. (2012) Ubc13 and COOH terminus of Hsp70-interacting protein (CHIP) are required for growth hormone receptor endocytosis. Journal of Biological Chemistry 287: 15533-15543.
- Zhang M, Windheim M, Roe SM, et al. (2005) Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 20: 525-538.
- Campbell SJ, Edwards RA, Leung CC, et al. (2012) Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. Journal of Biological Chemistry 287: 23900-23910.
- Motegi A, Liaw HJ, Lee KY, et al. (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci US A 105: 12411-12416.
- Motegi A, Sood R, Moinova H, et al. (2006) Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol 175: 703-708.
- Unk I, Hajdu I, Fatyol K, et al. (2006) Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA 103: 18107-18112.
- Juang YC, Landry MC, Sanches M, et al. (2012) OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell 45: 384-397.
- Nakada S, Tai I, Panier S, et al. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466: 941-946.
- Wiener R, DiBello AT, Lombardi PM, et al. (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nature Structural & Molecular Biology 20: 1033-1039.
- Wiener R, Zhang X, Wang T, et al. (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483: 618-622.
- Nikitina N, Bronner-Fraser M, Sauka-Spengler T (2009) The sea lamprey Petromyzon marinus: a model for evolutionary and developmental biology. Cold Spring Harb Protoc 1.
- Pancer Z, Saha NR, Kasamatsu J, et al. (2005) Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA 102: 9224-9229.
- Alder MN, Rogozin IB, Iyer LM, et al. (2005) Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310: 1970-1973.
- Pancer Z, Amemiya CT, Ehrhardt GR, et al. (2004) Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430: 174-180.
- Velikovsky CA, Deng L, Tasumi S, et al. (2009) Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nature Structural & Molecular Biology 16: 725-730.
- Van Wij SJ, Timmers HT (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J 24: 981-993.
- Cook WJ, Jeffrey LC, Sullivan ML, et al. (1992) Three-dimensional structure of a ubiquitin-conjugating enzyme (E2). Journal of Biological Chemistry 267: 15116-15121.
- Cook WJ, Jeffrey LC, Xu Y, et al. (1993) Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4. Biochemistry 32: 13809-13817.
- Cook WJ, Martin PD, Edwards BF, et al. (1997) Crystal structure of a class I ubiquitin conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry 36: 1621-1627.
Corresponding Author
Rigers Bakiu, Department of Aquaculture and Fisheries, Agricultural University of Tirana, Tirane, Albania.
Copyright
© 2017 Chen L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.