A Summary of Current Knowledge of Pandemic Coronaviruses Disease-19
Abstract
Experts around the globe named it Severe Acute Respiratory Syndrome Coronavirus 2, "SARS-CoV-2"; interchangeably it is called COVID-19 or coronavirus. COVID-19 is new emerged respiratory disease first identified in Wuhan city, China. It became pandemic which reaches around the world with a short time. It also has a significant crisis in human life, socio-economic activities. Currently, countries around the world become more interconnected in the economy and other human activities; because of this the crisis spread almost all countries and the crisis impact too. This review aimed to summarize the current knowledge of coronavirus. Therefore, this review was different topics about the current virus. The origin of the virus expected to be from animals, either from bat or pangolin or other unsuspected animals. Scientist identifies the taxonomical classification of these newly emerged pandemic viruses, accordingly, they suggest that it belongs to the same species with SARSr-CoV. In the meantime, in this review the following topics were addressed, the ways of transmission mainly based on contact with the infected person nasal droplets, the virus attachment and entry mechanism to the host cell, the clinical symptoms of the infected person, and the risk factors for this virus also another included topic. Additionally, human life and economic losses, prevention measures, promising treatments also comprised. As expected if the virus sourced from the animal, the human should avoid unnecessary contact with them. Moreover, the scientist should develop a methodology to prevent and cure for unexpected disease happening.
Keywords
Coronavirus, Covid-19, Pandemic, Bat/Pangolin, Risk factor, Treatment
Introduction
COVID-19 is a new respiratory virus that was first identified in the central Chinese city of Wuhan, in Hubei Province, in December 2019 [1]. It has since spread across China and around the world, with major outbreaks occurring in parts of Asia and Europe and the United States. On January 30, 2020, the WHO declared a Public Health Emergency of International Concern and on March 11 declared a pandemic, global outbreak of a disease [2]. According to the Oxford dictionary definition the name corona defined as (/kəˈrəʊnə/- a part of the body resembling or likened to a crown). The acronym COVID-19 stands for CO - corona', 'VI - viruses, 'D - disease','19 - the year 2019' [3].
Global experts on viruses have officially named the virus causing the outbreak "SARS-CoV-2". This is an abbreviation of "Severe Acute Respiratory Syndrome Coronavirus 2". After researching the new coronavirus, scientists determined it is closely related to the virus that caused the SARS epidemic in 2002 and 2003. The virus that caused SARS is known as SARS-CoV, so this new coronavirus is called SARS-CoV-2. They are common in many different species of animals, including camels, cattle, cats, and bats. Rarely, these animal coronaviruses can infect people and then spread between people [1].
Depending on the intensity of the pathogen, its mode of transmission, herd immunity, and prevalence and incidence of the illness and disease in the community disease outbreaks can be endemic when a disease is constantly present in a community. Disease outbreaks take the forms of pandemics when a large population, over a broad geographical area becomes susceptible to the disease agents, as a result of the simultaneous outbreaks of the diseases [4].
SARS-CoV-2 is the seventh coronavirus known to infect humans; SARS-CoV, MERS-CoV and SARS-CoV-2 can cause severe disease, whereas HKU1, NL63, OC43 and 229E are associated with mild symptoms [5]. Presently, there are four, endemic, coronavirus strains currently circulating in human populations (229E, HKU1, NL63, OC43). If the novel coronavirus follows the pattern of 2009 H1N1 pandemic influenza, it will also spread globally and become a fifth endemic coronavirus within the human population [6]. This review was aim to figure out major entity about COVID-19.
Expected Origin of COVID-19
Numerous mammal and avian species can be infected and the virus will cause acute disease. In several of these acute infections, the virus involved seems to have recently been adapted to the new host from other, often unknown sources. Various bat species have been found to support persistent asymptomatic infections by specific versions of SARS viruses. In fact six novel coronaviruses were isolated from six different bat species showing an astonishing diversity in bats [7].
Phylogenetic analysis indicates that all bat coronaviruses appear to have descended from a common ancestor. Only one of these bat groups includes SARS and SARS-like coronaviruses that adapted to acute human infections [7].
Huanan food market in Wuhan, was suggesting an intermediate animal host where adaptation to human transmission might have occurred. Live hedgehogs, badgers, snakes and turtledoves were sold at the Huanan market. Claims were made, but were not (yet) substantiated that snakes or pangolins were intermediate hosts for creating the coronavirus by recombination events [8].
It appears that most of the early cases had contact history with the original seafood market; however, the disease has now progressed to be transmitted by human-to-human contact. Full-length genome sequences were obtained from five patients at an early stage of the outbreak. The sequences are almost identical and share 79.6% sequence identity to SARS-CoV. Furthermore, 2019-nCoV is 96% identical at the whole-genome level to a bat coronavirus. Pair wise protein sequence analysis of seven conserved non-structural proteins domains show that this virus belongs to the species of SARSr-CoV. The close phylogenetic relationship to RaTG13 provides evidence that 2019-nCoV may have originated in bats [3].
Detailed understanding of how an animal virus jumped species boundaries to infect humans so productively will help in the prevention of future zoonotic events. Irrespective of the exact mechanisms by which SARS-CoV-2 originated via natural selection, the ongoing surveillance of pneumonia in humans and other animals is clearly of utmost importance [5].
Covid-19 is not a laboratory construct or a purposefully manipulated virus; instead, two scenarios that can plausibly explain the origin of SARS-CoV-2: (i) Natural selection in an animal host before zoonotic transfer; and (ii) Natural selection in humans following zoonotic transfer, whether selection during passage could have given rise to SARS-CoV-2 [5].
Not only bat, pangolin or other ambiguous reservoir hosts are suspected for various coronaviruses. Study showed that there was high species variety of coronavirus detected as potential pathogens of Malayan pangolins. Whole genome sequences of strains belonging to four coronavirus genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronaviruses) [9]. The genome sequence of the virus isolated pangolin-associated coronaviruses that belong to two sub-lineages of SARS-CoV-2-related coronaviruses, including one that exhibits strong similarity (99.83-99.92%) to SARS-CoV-2 in the receptor-binding domain. The discovery of multiple lineages of pangolin coronavirus and their similarity to SARS- CoV-2 suggests that pangolins should be considered as possible hosts in the emergence of novel coronaviruses [10]. Pangolin should be removed from wet markets to prevent zoonotic transmission [10].
Furthermore, another whole genome sequences study sagest, the Pangolin-CoV genome was highly similar throughout the genome to 2019-nCoV and Bat SARSr-CoV RaTG13, with sequence identity between 80 and 98%, except for the S gene. Pangolin-CoV is more similar to 2019-nCoV-like and Bat-CoV-RaTG13. The receptor-binding domain (RBD) of the S protein of Pangolin-CoV has only one amino acid difference from that of 2019-nCoV. One coronavirus isolated from Malayan pangolins showed 100%, 98.2%, 96.7% and 90.4% amino acid identity with 2019-nCoV in the E, M, N and S genes, respectively [11].
The comparison study from bat and pangolin by Li and his friends explain that BetaCoV/bat/Yunnan/RaTG13/2013 virus was more similar to the SARS-CoV-2 virus than the coronavirus obtained from the two pangolin samples (SRR10168377 and SRR10168378). Additionally, a unique peptide (PRRA) insertion was identified in the human SARS-CoV-2 virus, which may be involved in the proteolytic cleavage of the spike protein by cellular proteases, and thus could impact host range and transmissibility. Interestingly, the coronavirus carried by pangolins did not have the RRAR motif. Therefore, we concluded that the human SARS-CoV-2 virus, which is responsible for the recent outbreak of COVID-19, did not come directly from pangolins [12].
Taxonomy of COVID-19
Pairwise protein sequence analysis of seven conserved non-structural proteins domains show that this virus belongs to the species of SARSr-CoV [3]. Notably, 2019-nCoV was closely related (with 88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, collected in 2018 in Zhoushan, eastern China, but were more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%). Phylogenetic analysis revealed that 2019-nCoV fell within the subgenus Sarbecovirus of the genus Betacoronavirus, with a relatively long branch length to its closest relative bat-SL-CoVZC45 and bat-SL-CoVZXC21, and was genetically distinct from SARS-CoV. Notably, homology modeling revealed that 2019-nCoV had a similar receptor-binding domain structure to that of SARS-CoV, despite amino acid variation at some key residues [13].
A Blast search of the complete genomes of 2019-nCoV revealed that the most closely related viruses available on GenBank were bat-SL-CoVZC45 (sequence identity 87.99%; query coverage 99%) and another SARS-like betacoronavirus of bat origin, bat-SL-CoVZXC21 (accession number MG772934; 87.23%; query coverage 98%) [13]. The final genome of sequenced SARS-CoV-2 consists of a single, positive-stranded RNA that is 29,811 nucleotides long, broken down as follows: 8,903 (29.86%) adenosines, 5,482 (18.39%) cytosines, 5,852 (19.63%) guanines, and 9,574 (32.12%) thymines [14].
The virus genome consists of six major open-reading frames (ORFs) that are common to coronaviruses and a number of other accessory genes. Further analysis indicates that some of the 2019-nCoV genes shared less than 80% nucleotide sequence identity to SARS-CoV. The amino acid sequences of the seven conserved replicase domains in ORF1ab that were used for CoV species classification were 94.4% identical between 2019-nCoV and SARS-CoV, suggesting that the two viruses belong to the same species, SARSr-CoV [3].
The Scenario of Evolution of Virus
RNA and DNA viruses were thought to be evolutionary unrelated. However, the overall similarity between virus structures a protein coat enclosing a nucleoprotein filament at least suggests a common mechanism for their appearance. Three hypotheses have been pro- posed to explain the emergence of viruses: (i) They are relics of pre-cellular life forms; (ii) They are derived by reduction from unicellular organisms (via parasitic-driven evolution); (iii) They originated from fragments of genetic material that escaped from the control of the cell and became parasitic [15].
Ways of Transmission
The complete transmission routines of this virus among hosts are not yet know. It appears that the virus is becoming more transmissible between humans. Close monitor whether the virus continues to evolve to become more virulent [3].
Characteristics of COVID-19 more closely resemble those of influenza A (H1N1) pdm09, than those of SARS-CoV in 2003. However, prevention and control strategies are more difficult than those for influenza pandemic, since morbidity is different and no vaccine or specific treatment agents against this new virus are expected in the near future [16].
Common sense etiquettes will help mitigate the risk of transmission: hand hygiene, cough etiquette, and avoiding crowded places for high-risk people (e.g., elderly or immune compromised). People with mild symptoms of acute respiratory illness are not likely to benefit from medical care, regardless of etiologic agents. Visiting healthcare facilities without proper protection will risk others if one has COVID-19, or risk oneself otherwise. The public should be advised to stay home if mildly ill, and seek medical care in a coordinated way if symptoms persist or aggravate [16].
Airborne transmission was considered to be a possible explanation, other potential modes of transmission [17]. Although, cannot rule out other possible routes of transmission, as further investigation, including more patients, is required [3]. It appeared to be potentially from a zoonotic spillover at Huanan seafood wholesale market. Subsequent cases were because of person to person transmission [18].
Cases have been reported where a patient could infect their close contacts even after "apparent recovery" from the infection. Half of the patients continued to be viral positive even after the resolution of symptoms up to eight days. The viral clearance kinetics were 12 days [19].
Attachment and entry to the host
Many studies have shown that viruses use cellular proteins for multiple purposes in their replication cycles, including the attachment and entry into the cells, the initiation and regulation of RNA replication/transcription, the translation of their mRNAs, and the assembly of progeny virus [20]. COVID-2019 virus isolated from the broncho-alveolar lavage fluid of a critically ill patient could be neutralized by sera from several patients. Notably, the virus uses the same cell entry receptor angiotensin converting enzyme II (ACE2) as SARS-CoV [3].
Coronaviruses use their spike proteins to select and enter target cells and insights intonCoV-2019 spike (S)-driven entry might facilitate assessment of pandemic potential and reveal therapeutic targets. 2019-nCoV-S uses the SARS-coronavirus receptor, ACE2, for entry the cellular protease TMPRSS2 for 2019-nCoV-S priming. A TMPRSS2 inhibitor blocked entry and might constitute a treatment option [21].
Gene nomenclature Orf9b no. of amino acid residues in product (98), crystal structure of Orf9b, an alternative ORF within the N gene, may be involved in membrane attachment and associates with intracellular vesicles, consistent with a role in assembly of the virion [22].
ACE2 is known to be a cell receptor for SARS-CoV. To determine whether 2019-nCoV also uses ACE2 as a cellular entry receptor, virus infectivity studies conducted using HeLa cells that expressed or did not express ACE2 proteins from humans, Chinese horseshoe bats, civets, pigs and mice. Therefore, 2019-nCoV is able to use all ACE2 proteins, except for mouse ACE2, as an entry receptor to enter ACE2- expressing cells, but not cells that did not express ACE2, indicating that ACE2 is probably the cell receptor through which 2019-nCoV enters cells. However, 2019-nCoV does not use other coronavirus receptors, such as aminopeptidase N (APN) and dipeptidyl peptidase 4 (DPP4) [3].
Clinical Symptoms of COVID-19 Infected Person
According to [23], 1099 sampled laboratory-confirmed cases, found that the common clinical manifestations included fever (88.7%), cough (67.8%), fatigue (38.1%), sputum production (33.4%), shortness of breath (18.6%), sore throat (13.9%), and headache (13.6%). Moreover, from 34 patients the most common symptoms were fever (17 cases, 50%) and cough (13 cases, 38%). In the 34 cases, the white blood cell counts of 28 cases (82%) were normal. Five cases had white blood cell counts more than 10 × 109/L. One case had white blood cell counts less than 4 × 109/L [24].
In the severe patients, the neutrophil count, D-dimer, blood urea, and creatinine levels were higher significantly, and the lymphocyte counts continued to decrease. Additionally, inflammatory factors interleukin (IL)-6, IL-10, tumor necrosisfactor-α (TNF-α) increase, indicating the immune status of patients. The data showed that ICU patients had higher plasma levels of IL-2, IL-7, IL-10, granulocyte colony-stimulating factor (GCSF), 10 kD interferon- gamma-induced protein (IP-10), monocyte chemo- attractant protein-1 (MCP-1), macrophage inflammatory protein 1-α (MIP-1α), and TNF-α [23].
Another study total of 201 patients were included, the most commonly self-reported symptoms at onset of illness were fever (93.5%), cough (81.1%), productive cough (41.3%), dyspnea (39.8%), and fatigue or myalgia (32.3%). The majority (76.6%) of patients had fever with cough; (36.8%) had fever with dyspnea; (32.8%) had fever with fatigue, myalgia, or headache; and only (6.5%) presented with fever alone. About (95.0%) patients had findings of bilateral infiltrates on radiographic imaging, while 10 (5.0%) patients had unilateral infiltrates. Of the 201 patients, 165 (82.1%) required oxygen support in the hospital [25].
According to [23], from 41 patients, common symptoms at onset of illness were fever (98%), cough (76%), and myalgia or fatigue (44%); less common symptoms were sputum production (11 [28%] of 39), headache (three [8%] of 38), haemoptysis (two [5%] of 39), and diarrhoea (one [3%] of 38). Dyspnoea developed in 22 (55%) of 40 patients (median time from illness onset to dyspnoea 8.0 days [IQR 5.0-13.0]). 26 (63%) of 41 patients had lymphopenia. All 41 patients had pneumonia with abnormal findings on chest CT. Complications included acute respiratory distress syndrome (29%), RNAaemia (15%), acute cardiac injury (12%) and secondary infection (four 10%). (32%) patients were admitted to an ICU and six (15%) were died.
Risk Factors of COVID-19
It is difficult to determine the risk factors with the limited number of sample size, and the estimation leading to poor clinical outcomes has not been well delineated, it is early to determine the risk factor with small case for coronavirus [26].
In bivariate Cox regression analysis, risk factors associated with the development of ARDS (acute respiratory distress syndrome) and progression from ARDS to death included older age (hazard ratio [HR], 3.26; 95% CI 2.08-5.11; and HR, 6.17; 95% CI, 3.26-11.67, respectively), neutrophilia (HR, 1.14; 95% CI, 1.09-1.19; and HR, 1.08; 95% CI, 1.01-1.17, respectively), and organ and coagulation dysfunction (eg, higher lactate dehydrogenase [HR, 1.61; 95% CI, 1.44-1.79; and HR, 1.30; 95% CI, 1.11-1.52, respectively] and D-dimer [HR, 1.03; 95% CI, 1.01-1.04; and HR, 1.02; 95% CI, 1.01-1.04, respectively]). High fever (39 ℃) was associated with higher likelihood of ARDS development (HR, 1.77; 95% CI, 1.11-2.84) and lower likelihood of death (HR, 0.41; 95% CI, 0.21-0.82). Among patients with ARDS, treatment with methylprednisolone decreased the risk of death (HR, 0.38; 95% CI, 0.20-0.72). Older age was associated with greater risk of developing ARDS and death, likely because of less rigorous immune response. Although fever was associated with the development of ARDS, it was also associated with better outcomes. Several factors related to the development of ARDS were not associated with death, which indicates that different pathophysiological changes from hospital admission to development of ARDS and from development of ARDS to death may exist [25].
Additionally, ABO blood group showed different for coronavirus susceptibility. Meta-analyses two hospitals in China, the pooled data showed that blood group A had a significantly higher risk for COVID-19 (odds ratio-OR, 1.20; 95% confidence interval-CI 1.02 ~ 1.43, P = 0.02) compared with non-A blood groups, whereas blood group O had a significantly lower risk for the infectious disease (OR, 0.67; 95% CI 0.60 ~ 0.75, P < 0.001) compared with non-O blood groups. O blood group hospital staffs were less chance of getting infected. Anti-A antibodies specifically inhibited the adhesion of SARS-CoV S protein-expressing cells to ACE2-expressing cell lines. Given the nucleic acid sequence similarity and receptor angiotensin-converting enzyme 2 (ACE2) binding similarity between SARS-CoV and SARS-CoV-2. The lower susceptibility of blood group O and higher susceptibility of blood group A for COVID-19 could be linked to the presence of natural anti-blood group antibodies, particularly anti-A antibody, in the blood. This hypothesis will need direct studies to prove. There may also be other mechanisms underlying the ABO blood group-differentiated susceptibility for COVID-19 that require further studies to elucidate [27].
Another study on191 patients (135 from Jinyintan Hospital and 56 from Wuhan Pulmonary Hospital, in China), of whom 137 were discharged and 54 died in hospital. (48%) patients had comorbidity, with hypertension being the most common (30%) patients, followed by diabetes (19%) and coronary heart disease (8%). Multivariable regression showed increasing odds of in-hospital death associated with older age (odds ratio 1.10, 95% CI 1.03-1.17, per year increase; p = 0.0043), higher Sequential Organ Failure Assessment (SOFA) score (5.65, 2.61-12.23; p < 0.0001), and d-dimer greater than 1 μg/L (18.42, 2.64-128.55; p = 0.0033) on admission [28].
The human life loss due to this virus
According to WHO 30th March situation report, the total confirmed case up to the date of report comprehend 693,224 total confirmed case happen from which 33106 (4.77) were died this is not included the new confirmed cases, the percentage of mortality due to this disease might be higher only from this total confirmed cases. Table 1, indicates that the total disease and death case yet across the region of the world [29].
The Economic Loss
The world economy become drop, many factories closed or produce under their capacity, unemployment and deployment is expanded. The major human activities now restricted, the global business is significantly slowdown. Many flights are canceled even totally restriction, different countries election postponed and the world cup too. According to [29], the annual global GDP growth is projected to drop to 2.4% in 2020 as a whole, from an already weak 2.9% in 2019. Before picking up to around 3.5% in 2021 as the effects of the coronavirus fade and output gradually recovers. Relative to the SARS outbreak in 2003, the global economy has become substantially more interconnected, and China plays a far greater role in global output, trade, tourism and commodity markets.
The forecast, however, flagged the spread of the virus as a significant downside risk for the global and European economy. The direct impact through all channels is estimated to reduce real GDP growth in 2020 by 2.5% points compared to a situation where there would be no pandemic. Given that real GDP growth was forecast to be 1.4% for the EU in 2020, this would imply it could fall to just over -1% of GDP in 2020, with a substantial but not complete rebound in 2021[30]. Coronavirus new blow to economy growth expected to drop from 3.2% to 1.8% in Africa. Tourism experienced losses of up to $US7.2 billion and unanticipated increases in health spending of up to USD 10.6 billion, additionally, revenue losses could lead to unsustainable debt [31]. Additionally, for vaccine and treatment production and finding, the world will expense unanticipated amount of money.
Prevention Measure of COVID-19
Prevention of disease the first and invaluable measurement, how to prevent the disease before infected must be a big deal. According to [32] the following points were a measure suggested and followed by different population around the globe.
• Stay-at-home order to promote social distancing (at least 1.5 m) and control.
• Quarantining all travelers, including state residents, for 14 days.
• Having only essential workers go to their jobs, while others work from home if possible.
• Social gatherings are limited to 10 people or less.
• Bars Restaurants were closed except to provide food through drive-thru, take-out, or delivery.
• Theatres, entertainment centers, and visitor attractions closed.
• Residents are asked to avoid any non-essential travel.
• Services and activities at places of worship suspended.
• High-risk individuals are asked to stay home and take additional precautionary measures.
• People should not visit nursing homes, retirement facilities, or long-term care facilities.
• If someone household has tested positive for COVID-19, the entire household should stay home.
• Wash hands often with soap and water for at least 20 seconds, especially after going to the bathroom; before eating; and after blowing your nose, coughing, or sneezing.
• If soap and water are not readily available, use an alcohol-based hand sanitizer with at least 60% alcohol. Always wash hands with soap and water if hands are visibly dirty.
• Avoid touching your eyes, nose, and mouth with unwashed hands.
• Cover cough or sneeze with a tissue; then throw the tissue in the trash and use face mask.
• Clean and disinfect frequently touched objects and surfaces using a regular household cleaning spray or wipe.
Promising Treatments and Vaccines
Around the globe scientist are working on finding treatments and vaccine rapidly, unluckily proven treatment and vaccine is not yet developed. However, different findings suggest as an alternative drug which used before for other disease treatment until new treatment established. As 2019-nCoV is an emerging virus, an effective treatment has not been developed for disease resulting from this virus [1]. Double-blinded randomized clinical trials to determine the most effective treatments for COVID-19 are recommended. Accordingly, [23], suggest that treatment with methylprednisolone may be beneficial for patients who develop ARDS.
Another study confirms there was promising result from Chloroquine. Chloroquine is used to prevent and treat malaria and is efficacious as an anti-inflammatory agent for the treatment of rheumatoid arthritis and lupus erythematosus. The anti-viral and anti-inflammatory activities of chloroquine may account for its potent efficacy in treating patients with COVID-19 pneumonia. More than 100 patients have demonstrated that chloroquine phosphate is superior to the control treatment in inhibiting the exacerbation of pneumonia, improving lung imaging findings, promoting a virus-negative conversion, and shortening the disease course according to the news briefing [25].
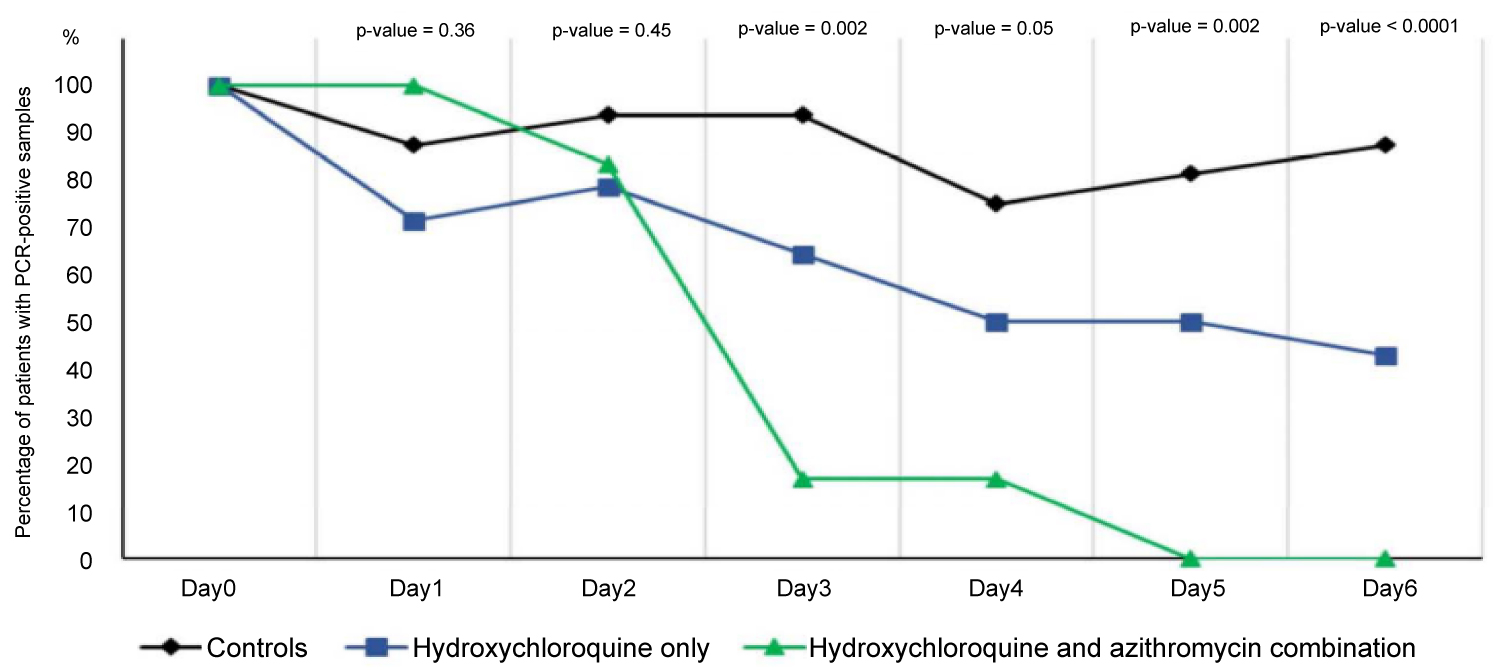
The French scientists trail the effect of hydroxychloroquine treatment as a single drug and the effect of hydroxychloroquine and azithromyc in combination. At day 6 post-inclusion, 100% of patients treated with hydroxychloroquine and azithromycin combination were virologicaly cured comparing with 57.1% in patients treated with hydroxychloroquine only, and 12.5% in the control group (p < 0.001). Hydroxychloroquine is efficient in clearing viral nasopharyngeal carriage of SARS-CoV-2 in COVID-19 patients in only three to six days [33]. However, according to [34], to the mean duration of viral shedding in patients suffering from COVID-19 in China was 20 days (even 37 days for the longest duration) (Figure 1) [34].
There is ample evidence of the in vitro and in vivo efficacy of CQ and its derivatives in the prevention and treatment of multiple species of coronavirus, including in vitro experiments with SARS-CoV-2 [28].
Ammonium chloride treatment strongly inhibited 2019-nCoV-S- and SARS-S-driven entry into TMPRSS2- 293T cells while inhibition of entry into TMPRSS2+ Caco-2 cells was less efficient, which would be compatible with 2019-nCoV-S priming byTMPRSS2 in Caco-2 cells [34]. According to Chaomin and his colleagues, among the patients with ARDS, of those who received methylprednisolone treatment, 23 of 50 (46.0%) patients died, while of those who did not receive methylprednisolone treatment, 21 of 34 (61.8%) died. The administration of methylprednisolone appears to have reduced the risk of death in patients with ARDS (HR, 0.38; 95% CI, 0.20-0.72; P = 0.003). Double-blinded randomized clinical trials to determine the most effective treatments for COVID-19 are still needed [35].
Currently, different countries around the world looking and tried experiments for herbal traditional medicine for treatments of COVID-19. Including China, Morocco, India and Ethiopia, however, yet only in China and India the experimental output is published. According to [21], in silico experiment of the natural compounds screened, 13 that exist in traditional Chinese medicines were also found to have potential anti-2019-nCoV activity. Further, 125 Chinese herbs were found to contain 2 or more of these 13 compounds. Of these 125 herbs, 26 are classically catalogued as treating viral respiratory infections [25]. In India according to [36] in-silico molecular docking analysis of the bio-active compounds found in the aqueous extract of Nilavembu kudineer chooranam against the ACE2 enzyme receptor which is the route of entry in the pathogenesis of Novel coronavirus [37].
Conclusion
Coronavirus becomes a pandemic disease within a short period due to the interconnected nature of the global population nowadays. The crisis is no some part of the world rather the whole world. Human life and economic loss continued, the current option for the virus is prevention only, until certified curative medicine found. Taking consideration the prevention measures must be the utmost action. People around the world must be prepared for the unknown anticipated risks, most government around the world not consider when it happens in China first until it comes its own country, even in many countries the measure start taken after the worsts happen. A definitive diagnosis should be made for the origin of the virus. If it is sourced from animals, avoid unnecessary contact with animals has an invaluable benefit. Additionally, if it is mutated, identification of the cause of the mutation is important to prevent the next unknown and unexpected pandemic disease emergency. The scientist should prepare the methodology of what should do if it is happening again.
Acknowledgements
For the scientific communities who availed their research output.
Fund
No funding available for this work.
Conflict of Interest
The author declared that there is no conflict of interest.
References
- CDC (2020) Centers for disease control and prevention, Cronovirus disease 2019 (Covid-19)- Frequently asked questions.
- Fan Wu, Su Zhao, Bin Yu, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265-269.
- Peng Zhou, Xing-Lou Yang, Xian-Guang Wang, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273.
- Rhyddhi Chakraborty (2015) Epidemics. Encyclopedia of Global Bioethics.
- Kristian G Andersen, Andrew Rambaut, W Ian Lipkin, et al. (2020) The proximal origin of SARS-CoV-2. Nature Medicine 26: 450-452.
- Ruiyun Li, Sen Pei, Bin Chen, et al. (2020) Substantial undocumented infection facilitates the rapid dissemination of Novel Coronavirus (COVID-19). Science 368: 489-493.
- Domingo E, Parrish CR, Holland JJ (2008) Origin and Evolution of Viruses. (2nd edn ), Elsevier.
- Harald Brüssow (2020) The Novel Coronavirus - A snapshot of current knowledge. Microb Biotechnol 13: 607-612.
- Ping Liu, Wu Chen, Jin-Ping Chen (2019) Viral metagenomics revealed sendai virus and coronavirus infection of malayan pangolins (Manis Javanica). Viruses 11: 979.
- Tommy Tsan-Yuk Lam, Na Jia, Ya-Wei Zhang (2020) Identifying SARS-CoV-2 related coronaviruses in malayan pangolins. Nature 583: 282-285.
- Kangpeng Xiao, Junqiong Zhai, Yaoyu Feng, et al. (2020) Isolation and characterization of 2019-NCoV-like coronavirus from malayan pangolins. BioRxiv.
- Xingguang Li, Junjie Zai, Qiang Zhao, et al. (2020) Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol 92: 602-611.
- Roujian Lu, Xiang Zhao, Juan Li, et al. (2020) Genomic characterisation and epidemiology of 2019 Novel Coronavirus: Implications for virus origins and receptor binding. The Lancet 395: 565-574.
- Sah R, Rodriguez-Morales AJ, Jha R, et al. (2020) Crossm complete genome sequence of a microcystin-degrading. Microbiology Resource Announcements 5: 4-5.
- Forterre Patrick (2006) The origin of viruses and their possible roles in major evolutionary transitions. Virus Research 117: 5-16.
- Kyungmin Huh, Hyoung Shik Shin, Kyong Ran Peck (2020) Emergent strategies for the next phase of COVID-19. Infect Chemother 52: 105-109.
- Vincent CC Cheng, Susanna KP Lau, Patrick CY Woo, et al. (2007) Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clinical Microbiology Reviews 20: 660-694.
- Ian Sinha (2020) Guidance for the clinical management of children admitted to hospital with suspected COVID-19. British Pediatrics Respiratory Society.
- Chang De, Guoxin Mo, Xin Yuan, et al. (2020) Time kinetics of viral clearance and resolution of symptoms in Novel Coronavirus infection. American Journal of Respiratory and Critical Care Medicine 201: 1-12.
- Arber W (1977) Current topics in microbiology and immunology, immunological communications.
- Markus Hoffmann, Hannah Kleine-Weber, Nadine Krüger, et al. (2020) The Novel Coronavirus 2019 (2019-NCoV) uses the SARS-Coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv.
- Christoph Meier, A Radu Aricescu, Rene Assenberg, et al. (2006) The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure 14: 1157-1165.
- Chaolin Huang, Yeming Wang, Xingwang Li, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395: 497-506.
- XF Wang, J Yuan, YJ Zheng, et al. (2020) Retracted: Clinical and epidemiological characteristics of 34 children with 2019 novel coronavirus infection in Shenzhen. Zhonghua Er Ke Za Zhi 58: E008.
- Chaomin Wu, Xiaoyan Chen, Yanping Cai, et al. (2020) Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China, JAMA Intern Med 180: 1-11.
- Jin-Nong Zhang, Wen-Jing Wang, Bo Peng, et al. (2020) Potential of Arbidol for post-exposure prophylaxis of COVID-19 transmission: A preliminary report of a retrospective case-control study. Curr Med Sci 40: 480-485.
- Jiao Zhao, Yan Yang, Hanping Huang, et al. (2020) Relationship between the ABO blood group and the COVID-19 susceptibility. MedRxiv.
- Fei Zhou, Ting Yu, Ronghui Du, et al. (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. The Lancet 395: 1054-1062.
- WHO Situation Report (2020) Coronavirus Disease 2019 (COVID-19) Situation Report - 70. DroneEmprit.
- OECD (2020) Interim economic assessment, coronavirus: The World Economy at Risk.
- European Commission (2020) Coordinated economic response to the COVID-19 outbreak.
- UNECA (2020) Economic Commission for Africa, Economic Impact of the COVID-19 on Africa.
- Jianjun Gao, Zhenxue Tian, Xu Yang (2020) Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14: 72-73.
- Philippe Gautret, Jean-Christophe Lagier, Philippe Parola, et al. (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open- label non-randomized clinical trial. Int J Antimicrob Agents 56: 105949.
- John Kearney (2020) Chloroquine as a potential treatment and prevention measure for the 2019 Novel Coronavirus: A review. Journal of Chemical Information and Modeling 53: 1689-1699.
- Deng-hai Zhang, Kun-lun Wu, Xue Zhang, et al. (2020) In silico screening of chinese herbal medicines with the potential to directly inhibit 2019 Novel Coronavirus. Journal of Integrative Medicine 18: 152-158.
- M Walter, Thomas, C Samuel Justinraj, et al. (2020) Effect of nilavembu kudineer in the prevention and management of COVID - 19 by inhibiting ACE2 receptor. Siddha Papers.
Corresponding Author
Fangyuan Chen, School of Mathematical Sciences, Beijing Normal University, Beijing, 100875, China.
Copyright
© 2020 Alebachew GW. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.