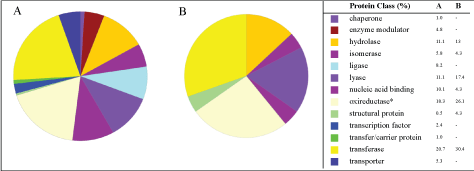Isolated Pseudomonas Aeruginosa from Mining Site: Proteome Changes Due to Presence of Copper
Abstract
The copper mining generates high amounts of residues. Thus, these residues and mining sites need to be treated correctly to avoid environmental risks. The identification of intracellular proteins provides remarkable information on microbial physiology that may contribute to the understanding of bioremediation (copper) by microorganisms. Thus, the aim of this study was to evaluate the effect (bactericide and bacteriostatic) of copper on Pseudomonas aeruginosa growth (isolated from mining sites) and then identify changes in the microbial proteome under different stress conditions induced by copper. Samples were collected (after 32 h of fermentation), centrifuged, washed and digested by trypsin. The peptides were analyzed using nano LC-ESI-Q-TOF (PepMap-15 cm × 75 µm column under gradient mode from 2 to 98% (v/v) of acetonitrile 0.1% formic acid for 180 minutes). MS precursors and MS/MS products were acquired over a 50-3000 m/z range. The data were processed via Protein Scape and classified according to the Panther system. The minimum inhibitory concentration (copper) was between 4-4.5 mM, in which bacteriostatic effects between 4.5-6.5 mM and bactericidal above 7 mM were observed. Nano LC-ESI-Q-TOF system; Protein Scape (Bruker-Daltonics) and Panther classification system composed one the best tools for proteomic analysis (bioanalytical chemistry). The concentration of 3 mM of copper was chosen based on obtained MIC data. The number of protein classes expressed by Pseudomonas aeruginosa was inversely proportional to the concentration of copper: 13 and 7 protein classes to 0 and 3 mM of copper, respectively demonstrating the metabolic stress. These results are aligned to MIC and MBC data. In this sense, proteins related to energy metabolism-TCA cycle and Pyruvate-were (bio)synthesized in both experiments, which proved the cell viability (indispensable for bioremediation).
Keywords
Copper mining, Pseudomonas aeruginosa, Proteomics
Introduction
Mining is one of the most environmentally hazardous processes (anthropogenic activities), since it generates large amounts of waste [1]. In this sense, the extraction of copper, particularly in Brazilian mining fields, significantly increased in 2004 due to the start up of Sossego (copper exploration mine) [2,3] . In 1998, 34,000 tons of copper were extracted; and this number increased to 216,000 tons in 2008. In addition, it was estimated that in 2030 the copper extraction will reach ≈ 374,000 tons [3].
In general, the residues of copper extraction (mining) have high content of sand (percolation), making harder to do the treatment and disposal correctly [2]. Therefore, chemical and physical remediation technologies, generally, show low efficiency and high cost [1,2]. Thus, the remediation by microorganisms such as bacteria, yeasts, filamentous fungi, algae and plants (bioremediation) appears as an interesting alternative to correctly treat these types of residues and also present potential for metal recovery (residues) [1,2,4].
The ability to absorb/complex metals by microorganisms are known as biosorption/bioaccumulation which can be used for bioremediation of toxic metal ions. The biosorption/bioaccumulation of metals by microorganisms involves physical-chemical processes such as adsorption, ion exchange, chelation, microprecipitation, etc [1].
Regarding the microbial biosorption of metals, the use of genetic engineering techniques in cell wall has been investigated. Changes in the cell wall aim to increase the number of amino acids (mainly ionics) [1,4-6]. In this context, the relationship between the bioaccumulation of copper and microorganisms is, intrinsically, related to the understanding of copper as an active compound in the cell metabolism, for instance metallothionein and chaperone proteins.
Proteomics is one of the most recent techniques used for comparison among different cellular physiologies. Microorganisms can be used to (bio)remediate contaminated soils or waters. These contaminated soils or waters can be used as culture medium (bioprocesses), in which contaminants such as copper will be absorbed by microorganism. Therefore, the identification of intracellular proteins could provide remarkable information on microbiological physiology, which may lead to a better understanding of copper bioremediation by microorganisms. The proteomic data of Pseudomonas aeruginosa (isolated from mining sites) using nanoLC-ESI-Q-TOF system allowed identifying proteome changes due to the concentration of copper (stress conditions).
Material and Methods
Cell growth (inoculum)
The bacterium isolated from mining site, was inoculated on Brain Heart Infusion (BHI) agar and incubated at 37 ℃ for 24 h. The microbial culture was transferred to 4 mL of 0.85% saline solution (initial microbial solution). Then, 2 mL of homogenized culture were removed for adjusting the optical density in spectrophotometer values between 0.08 and 0.1 at 625 nm, equivalent to 0.5 turbidity on the McFarland scale, or 1 to 2 × 108 CFU/mL. Then, dilutions were made in Mueller-Hinton or BHI broth to reach the concentration of 1 × 106 CFU/Ml [7].
Minimum inhibitory concentration and minimal bacteriostatic concentration
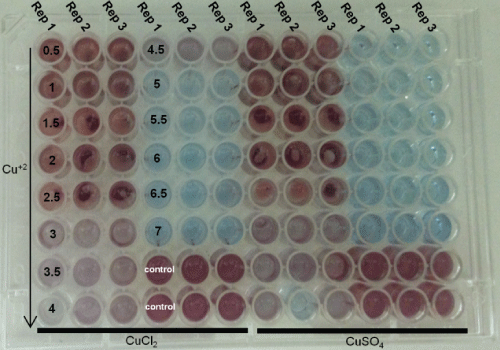
The Minimum Inhibitory Concentration (MIC) was defined as the lowest concentration that prevents (visibly) the microbial growth (Figure 1). The antimicrobial activity was carried out by microdilution, in which 0.1 mL of broth BHI was added to each well ELISA microplate (Spectra Max 340, Molecular Devices Ltd.) following by a serial dilution of 0.1 mL of the samples (biomass-32 h fermentation). Then, 0.1 mL of standardized inoculum was added to each well, resulting in a final concentration of 5 × 105 CFU/mL. The tests were developed simultaneously for the negative control (culture medium only), positive control (culture medium + microorganism) and sterility control (culture medium + sample). The microplates were incubated at 37 ℃ during 24 h. Then, 25 μL of 0.1% triphenyl tetrazolium chloride (indicator) was added and incubated again at 37 ℃ during 2 h. Whereas the Minimal Bacteriostatic Concentration (MBC) was performed using 5 μL from each well (ELISA microplate) in which, after inoculation in BHI agar for 24 h at 30 ℃, the microbial growth was observed (+ bacteriostatic or-bactericidal) [7].
Proteomic methodology
Sample biomass in absence and presence of 3 mM cooper ions were collected (fermentation 32 h, 30 ℃), centrifuged and washed in saline solution 0.85%. Thus, the further analysis used only intracellular proteins (Pseudomonas aeruginosa biomass). The biomass was then digested by trypsin (18 h, 37 ℃) producing peptides which allow identifying the intracellular proteins of Pseudomonas aeruginosa. Peptides were separated by NanoLC chromatography and then identified by ESI-Q-TOF, that is NanoLC-ESI-Q-TOF system (Impact-II-Bruker-Daltonics) containing Pep Map column (15 cm × 75 μm; Thermo-Scientific) by using a gradient from 2 to 98% (v/v) of acetonitrile 0.1% formic acid during 180 min. The instrument was operated in positive mode and MS spectra were acquired at spectra rate 2 Hz. MS precursors and MS/MS products were acquired over a 50-3000 m/z range. The data were processed by Protein Scape version 3.1.5 (Bruker-Daltonics), which is a bioinformatics platform with MASCOT database search engine to perform MS/MS data search against the SwissProt databases to identify proteins, in which the following search parameters were used: (1) Taxonomy: All Entries; (2) Enzyme: Trypsin; (3) Maximum missed cleavages: 1; (4) Fixed modification: Carbamidomethyl (C); (5) Variable modification: Oxidation (O); (6) Peptide tolerance: ± 0.02 Da; (7) MS/MS tolerance: ± 0.05 Da; (8) Significance Threshold p < 0.05); (9) Ions Score Cut-off: 15; (10) Protein and Peptide Assessment-If Mascot Score > 13.0 for both. Panther classification system was used to analyze all identified proteins through the specifically selecting Pseudomonas aeruginosa organism and then following by ontologies such as protein class, molecular function and biological process.
Results and Discussion
Minimum inhibitory concentrations: Bactericide and bacteriostatic effects
The MIC was defined as the lowest concentration in which there was no red color appearance (microbial growth indicator) [4,7,8]. MIC and MBC tests were carried out by using standard solutions of CuCl2 and CuSO4 (Table 1).
The Figure 1 and Figure 2 show the results obtained for the determination of MIC, in which the more reddish staining indicates higher microbial growth (Figure 1). Thus, the analysis of results indicated a subtle difference between CuCl2 and CuSO4, in which the MIC for CuCl2 was between 4.5 (light red) and 5 mM (blue) of copper ions, whereas the MIC for CuSO4 was between 4 (light red) and 4.5 mM (blue) of copper ions.
Similar approach (Figure 1) was carried out by Elguindi, et al. [9], who investigated the MIC (copper ions) for several Pseudomonas aeruginosa strains. The authors reported that the MIC was 2 mM, copper ions (CuCl2 H2O). However, the MIC was calculated when 50% growth inhibition was observed.
When compared to data found by Elguindi, et al. [9], the MIC results showed in Figure 1 are higher, very likely due to the adaptation of the strain isolated in copper mine or/and because the methodology used in this study considered the MIC when ≈ 100% growth inhibition was observed.
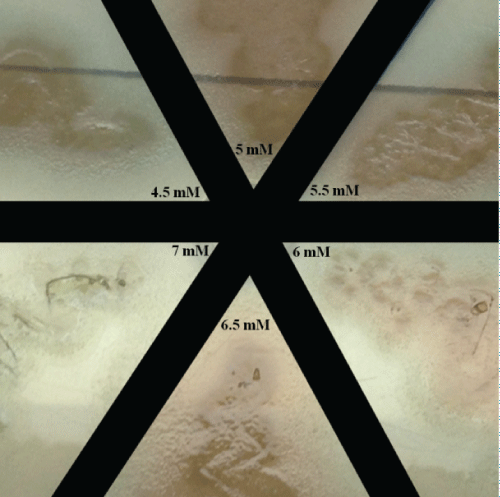
As already mentioned, the blue coloration (Figure 1) indicates that Pseudomonas aeruginosa did not grow, which means bacteriostatic or bactericidal effects had occurred. Thus, the media (blue wells-Figure 1) were used as inoculum in Petri dish, in which the growth on Petri dish demonstrates bacteriostatic effect.
The bacteriostatic effect was observed between 4.5 and 6.5 mM of copper ions, whereas at 7 mM bactericidal effect was found (Figure 2).
Cell growth
In our previous experiments we have identified that the use of culture medium described by [10] did not provide enough nutrients for the Pseudomonas aeruginosa cultivation isolated from mining site, since the glucose content did not reach ≈0 g/L after 24 hours of cultivation (data not shown). Therefore, experiments modifying the composition of the medium were tested. In this context, the addition of 2 g/L of urea (CH4N2O) to the medium became possible to cultivate P. aeruginosa. Modified synthetic medium for Pseudomonas aeruginosa growth is shown in Table 2.
Proteomic analysis: Changes induced by copper addiction
Gene Ontology (GO) is a major bioinformatics tool to related gene and gene product attributes across all species, which can be divided in molecular function, biological process, cellular component, protein class and pathways. In this study, the analyses of identified proteins were performed by PANTHER-Protein Analysis Though Evolutionary Relationships-classification system [11].
Protein class
The number of protein classes expressed by Pseudomonas aeruginosa was inversely proportional to copper ions concentration; in which 13 and 7 protein classes were found, for 0 and 3 mM of copper ions, respectively, which implies in metabolic stress mediated by copper. Chaperone, enzyme modulator, ligase, transcription factor, transfer/carrier protein and transporter were no longer expressed in the experiments with 3 mM of copper ions as show Figure 3A and Figure 3B. In addition, the genes and protein class hits (data no shown) were significantly higher in experiments without copper (266 and 208), when compared to the experiments with 3 mM of copper in the culture media (34 and 23). Thus, it is clear that the presence of cooper at 3 mM of copper affected the proteomics of Pseudomonas aeruginosa (Figure 3).
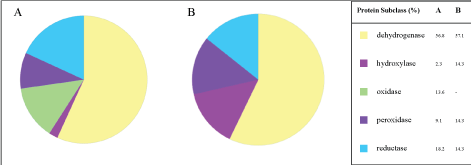
Oxidoreductases are enzymes that catalyze oxidation-reduction reactions. As illustrated in the Figure 3, in both experiments the oxireductase class was the most expressed protein class (% of gene hit against total protein class) - 18.3% and 26.1%; 0 mM and 3 mM, respectively. Even though these differences (Figure 3), The GO level 1 of the Protein Class (Figure 4) showed a much more slight difference, that is, similar % of dehydrogenase 56.8 and 57.1% and reductase 18.2 and 14.3%; 0 mM and 3 mM, respectively.
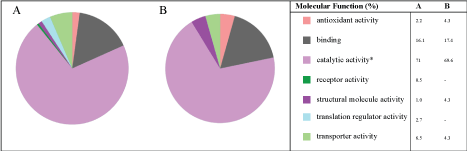
Molecular function
In GO, molecular function details reactions at molecular level, for instance binding, catalytic, antioxidant or transporter activities. The analysis of data showed that 7 classes of proteins (molecular function) were expressed at zero concentration of copper (Figure 5A). On the other hand, 5 classes of proteins were identified in the experiments with 3 mM of copper ions, in which the following classes of proteins: Translation regulator activity and receptor activity were no longer expressed in the experiments with 3 mM of copper (Figure 5B). In addition, in both experiments, proteins with catalytic activity (enzymes) were mostly expressed ≈ 70%.
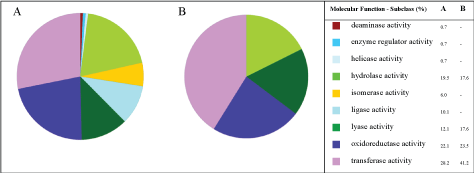
It can be observed that the number of proteins with catalytic activity reduced from 9 to 4, in the cultures without copper and with 3 mM of copper ions-Figure 6A and Figure 6B, respectively. Furthermore, in both experiments, proteins such as hydrolase activity, oxidoreductase activity and transfer activity were the major groups of (bio)synthesized proteins.
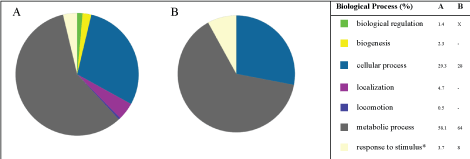
Biological process
Compared to molecular function, biological process relates a series of events (more than one distinct step) that are carried out by assemblies of molecular functions, for instance biological regulation, cellular components organization, metabolic process or response to stimulus. The biological process proteins were expressed in both cultivations, in absence and presence of copper (Figure 7A and Figure 7B).
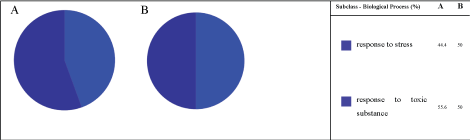
It is worth noting that even in the absence of copper, response to stress proteins (8%) were identified (Figure 8), which means that copper ions were not the only responsible for stressing the Pseudomonas aeruginosa; metabolites, other compounds of culture medium, etc, can stress it. In addition, even with the increased on expression of response to stress proteins, as shows Figure 7A → Figure 7B, 3.7 to 8%, respectively; the subsets are quite similar (Figure 8), that is, composed only by response to stress proteins and response to toxic substance proteins-approximately 50% each.
Pseudomonas aeruginosa bacteria in absence (0 mM) and presence of 3 mM of copper ions, A and B, respectively.
Pathways
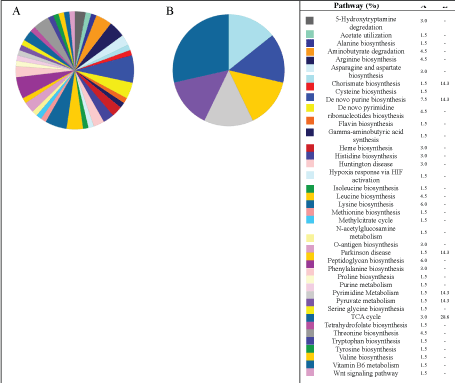
As illustrated in Figure 9, the pathways are the most diversified GO data, in which the most of pathways were suppressed by the presence of ions copper. Whereas in the experiments without copper, several amino acids pathways were identified such as arginine, alanine, asparagine biosynthesis among other. In this sense, it is worth noting that both experiments presented the TCA cycle and Pyruvate metabolism proteins, that are related to central energy metabolisms. Some pathways expressed in both experiments are illustrated Supplementary Figures.
Conclusion
NanoLC-ESI-Q-TOF; ProteinScape (Bruker-Daltonics) through MASCOT engine search and protein database SwissProt; and Panther classification system composed one the best tools for proteomic analysis (bioanalytical chemistry) Supplementary Data. The concentration of 3 mM of copper was chosen based on obtained MIC data in which was observed Pseudomonas aeruginosa cell growth (protein synthesis) that allowed identifying significant proteomic changes induced by copper. When compared to the experiment in absence of copper, the experiment with 3 mM of copper showed lower number of proteins-for all ontologies (molecular function, biological process, etc). These results are aligned to MIC and MBC data (↑ copper ≈ ↓ microbial growth ≈ ↓ protein synthesis). In this sense, proteins related to energy metabolism-TCA cycle and Pyruvate-(fundamental metabolism) were (bio)synthesized in both experiments, which proved the cell viability (indispensable for bioremediation).
Acknowledgments
The São Paulo Research Foundation-FAPESP (Project number 2013/50218-2) and BNDES-FUNTEC/VALE (Project number 2542).
References
- Kotrba P, Mackova M, Macek T (2011) Microbial biosorption of metals. Springer, Dordrecht Heidelberg, London, New York.
- Andreazza R (2009) Potencial do uso de bacterias e plantas para a remediacao de cobre em areas de vitivinicultura e de rejeito de mineracao de cobre no Rio Grande do Sul. Federal University of Rio Grande do Sul.
- (2016) Departamento Nacional de Producao Mineral.
- Salvadori MR, Ando RA, Nascimento CAO, et al. (2014) Intracellular biosynthesis and removal of copper nanoparticles by dead biomass of yeast isolated from the wastewater of a mine in the brazilian Amazonia. PLoS One 9: e87968.
- Cooksey DA, Azad HR (1992) Accumulation of copper and other metals by copper-resistant plant-pathogenic and saprophytic Pseudomonads. Appl Environ Microbiol 58: 274-278.
- Colin VL, Villegas LB, Abate CM (2012) Indigenous microorganisms as potential bioremediators for environments contaminated with heavy metals. Int Biodeter Biodegr 69: 28-37.
- Simiqueli APR (2014) Producao continua de surfactina utilizando manipueira como substrato e estudo de sua aplicacao em conjunto com oleos essenciais. University of Campinas.
- Sabry SA, Ghozlan HA, Abou-Zeid DM (1997) Metal tolerance and antibiotic resistance patterns of a bacterial population isolated from sea water. J Appl Microbiol 82: 245-252.
- Elguindi J, Wagner J, Rensing C (2009) Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J Appl Microbiol 106: 1448-1455.
- Atlas RM (2010) Handbook of microbiological media. (4th edn), CRC Press, Taylor & Francis Group 600 Broken Sound Parkway NW, Boca Raton, Florida 895.
- Mi H, Huang X, Muruganujan A, et al. (2016) PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: 183-189.
Corresponding Author
Cristiano J Andrade, Dempster MS Lab, Department of Chemical Engineering, Polytechnic School, University of São Paulo, Escola Politécnica, São Paulo-SP, Brazil, Zip Code: 05350-000, Tel: +55-19-98154-3393.
Copyright
© 2017 Andrade CJ, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.